1000/1000
Hot
Most Recent

Scedosporium/Lomentospora cell wall components, including peptidorhamnomannans (PRMs), α-glucans and glucosylceramides, are important immune response activators following their recognition by TLR2, TLR4 and Dectin-1 and through receptors that are yet unknown. After recognition, cytokine synthesis and antifungal activity of different phagocytes and epithelial cells is species-specific, highlighting the poor response by microglial cells against L. prolificans. Moreover, a great number of Scedosporium/Lomentospora antigens have been identified, most notably catalase, PRM and Hsp70 for their potential medical applicability. Against host immune response, these fungi contain evasion mechanisms, inducing host non-protective response, masking fungal molecular patterns, destructing host defense proteins and decreasing oxidative killing.
Among microbial pathogens, fungi have historically been treated as a minor threat to human health compared to other infectious agents such as viruses or bacteria, because they affected a lower percentage of patients and antifungal treatments were effective for most of them. In recent decades, however, the outlook has been changing radically, as fungal infections increase in frequency as a result of a substantial increase in immunosuppressive infections, such as those generated by the human immunodeficiency virus (HIV), as well as in the use of more potent immunosuppressive drugs and in invasive medical interventions. Moreover, resistance to drugs has become more and more common, which may be due to several reasons, including the use of environmental fungicides as reported in Aspergillus resistance to azole [1].
Even amongst pathogenic fungi, there has been a discriminatory behavior in favor of genera such as Candida, Cryptococcus or Aspergillus, because of the higher incidence of infections they cause compared to other fungi [2][3][4][5]. However, rare mycoses, especially those caused by filamentous fungi, are on the rise [5], probably as a result of the use of antifungal prophylaxis in all patients at risk of opportunistic fungal infection. This routine has proven effectivity in preventing the most frequent molds [6] but may be contributing to the emergence of less common mold infections such as those caused by the Lomentospora and Scedosporium species, which are highly resistant to current antifungal drugs [7][8].
The species of the genera Scedosporium has been reclassified in recent years. The genus currently includes 10 species [9][10][11] formerly classified within the Pseudallescheria/Scedosporium complex; the dual name was replaced by the genus Scedosporium following the “Amsterdam Declaration on Fungal Nomenclature”, which recommends a single-name system for all fungi [12]. Some years later, the species Scedosporium prolificans was separated and redefined as Lomentospora prolificans [13].
The Scedosporium/Lomentospora species are saprophytes usually found in soil, especially from human-disturbed ecosystems [14], but can also behave as opportunistic pathogens. They are considered as emerging fungal pathogens due to the increasing frequency of infections associated with high mortality rates. These infections appear mainly as a consequence of direct inoculation, respiration, and aspiration of polluted water during near-drowning events, with a remarkable prevalence after tsunami catastrophes [9][11][15][16]. In any case, once the infection has disseminated, systemic infections lead to a worse scenario. They also show neurotropism, meaning that central nervous system (CNS) infections are a probable consequence [9][17].
Specifically, they can cause a wide variety of diseases from localized infections in immunocompetent individuals to disseminated infections in those that are immunocompromised [14]. Within the immunocompromised group, transplant recipients and patients with primary hematological malignancies are among those at highest risk of suffering from these mycoses [9][11][15][18][19]. In the case of immunocompetent individuals, those who suffer from cystic fibrosis (CF) are susceptible to chronic airway colonization. In these patients, Scedosporium/Lomentospora species show a prevalence ranging between 0–25%, ranking second among filamentous fungi, after Aspergillus fumigatus [9][11][15][20][21][22][23][24]. This colonization is considered a risk factor for the development of an infection [24] and contributes to chronic inflammation, which can lead to the progressive deterioration of the lung function, as demonstrated for A. fumigatus [24][25][26].
Diagnosis is still based on culture-dependent methods and serological tests are only performed on a “home-made” basis, [27][28], but are not commercially available. This makes it impossible to differentiate between an airway colonization and a respiratory infection. Treatment is difficult, as these fungi show very limited susceptibility to current antifungals. European guidelines currently recommend voriconazole as a first-line treatment, together with surgical debridement when possible. In some cases, combinations of antifungals are needed, of which the most studied are voriconazole and terbinafine [9][29][30][31][32]. However, combination therapy is only supported by some reports and is therefore only moderately recommended.
As in most microorganisms, activation of an innate immune response to fungal infections is induced by the recognition of molecular components shared by many fungal species, found mainly on the cell wall, known as Pathogen Associated Molecular Patterns (PAMPs). To achieve this, the cells in the immune system use Pattern Recognition Receptors (PRRs) which recognize microbial molecules and activate signaling pathways that generate induction of the synthesis of pro-inflammatory cytokines, phagocytosis and adaptive immunity [33][34], as described below. The PRRs most implicated in the recognition of fungal pathogens are usually Toll-like receptors (TLRs), C-type Lectin Receptors (CLRs), Dectin-1 and 2, and Mannose Receptor/CD206 (MR) [33]. Specifically, the recognition of Scedosporium/Lomentospora has not yet been described in depth, but some of these PRRs have been indicated as being of paramount importance (Figure 1 and Figure 2).
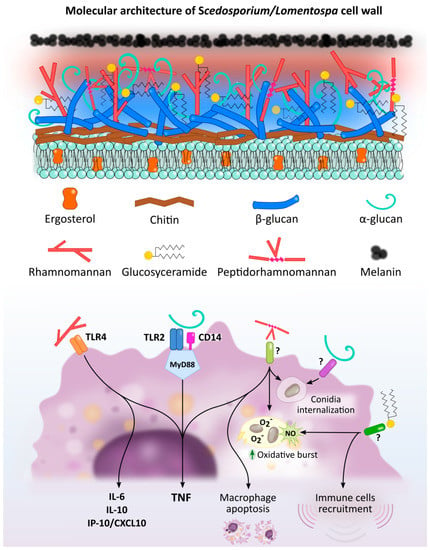
Figure 1. Scedosporium/Lomentospora cell wall components induced macrophage activation and Pattern Recognition Receptors (PRR) involved in these interactions. Rhamnomannans binds to TLR4 stimulating TNF, IL-6, IL-10 and IP-10/CXCL10 release. α-glucans stimulates TNF release through TLR2, CD14 and MyD88. It is also involved in conidia uptake, but the receptor involved in this process is unknown. Peptidorhamnomannan (PRM) is involved in fungal phagocytosis, induces macrophages death, and stimulates production of TNF, nitric oxide (NO) and superoxide (O2−). Glucosylceramide (GlcCer) induces NO and superoxide O2− production, increases fungal conidia killing, and promotes the recruitment of immune cells, including macrophages. The GlcCer and PRM receptor/s implicated in this activation have not been elucidated.
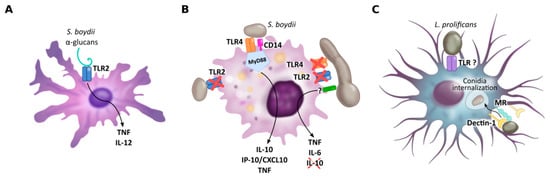
Figure 2. PRRs of dendritic cells (A), macrophages (B) and microglial cells (C) implicated in Scedosporium/Lomentospora recognition. Dendritic cell TLR2 is necessary for α-glucans recognition and consequent IL-12 and TNF secretion. Macrophage TLR4, together with CD14 and MyD88, but not TLR2, is required to induce macrophage activation by conidia, whereas hyphae recognition is independent of TLR2 and TLR4. Microglial cell mannose receptors, but especially Dectin-1, mediate L. prolificans conidia uptake. Other receptors may be involved in this process.
Recognition of fungal molecules leads to an activation of immune cells to respond against the microbial presence. It is obvious that the pathways activated are different depending on the fungal molecule recognized and the receptor used. However, the type of immune cell is also a determining factor. The following paragraphs summarize current knowledge on the response given by monocyte-derived macrophages, peritoneal macrophages, neutrophils, microglial cells, and epithelial cells against Scedosporium/Lomentospora (Figure 3).
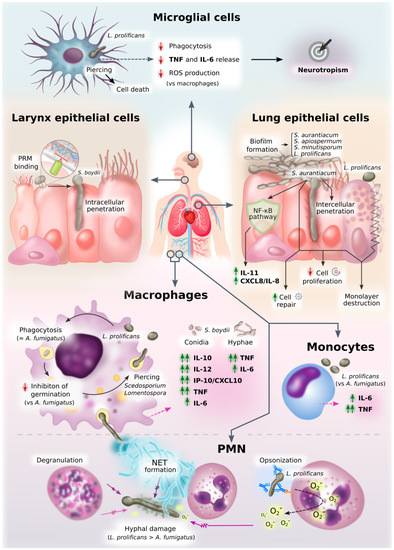
Figure 3. Diagram of innate immune response against Scedosporium/Lomentospora. This figure graphically represents an overview of all the information on innate cell interactions with Scedosporium/Lomentospora gathered here.
Antigen-presenting cells—mainly dendritic cells—digest the potential antigens and present them to T cells, promoting differentiation of T-helper (Th), cytotoxic (Tc) and regulatory (Treg) cells depending on the stimulus and PRR involved. Specifically, to overcome fungal infections Th1, Th2 and Th17 cells are of utmost importance [9][35]. For example, In the case of the widely studied fungal pathogen C. albicans, Th1 and Th17 responses have been shown to be protective, whereas Th2 type response is related to a worse infection outcome [36]. However, in the case of Scedosporium/Lomentospora infections, there are almost no studies regarding T cells, and the data presented are not enough to establish the role of T cells in these infections. In a study conducted among HIV patients, Tammer et al. (2011) found that patients with proven invasive scedosporiosis presented lower CD4+ cell levels than patients that were only colonized [37]. However, the T cells are not the only CD4+ cells, and thus a direct correlation between invasive scedosporiosis and low T cell counts cannot be established. In addition, in mice infections conducted by Xisto et al. (2016), they found that Th17 levels did not significantly change when mice were infected with L. prolificans, although T memory cell populations did significantly increase, probably as a result of the ongoing infection [38]. Finally, another study showed that L. prolificans lysates were able to expand specific T cells for this fungus in vitro, mainly CD4+ cells (81% CD4+ vs. 9.5% CD8+) [39]. Taken together, these three studies might indicate that a T-cell response is activated against Scedosporium/Lomentospora infections, primarily a Th response other than Th17, but many more studies need to be conducted to elucidate fully the interaction of T cells with this species complex.
An essential role of Th cells, in this case specifically of Thf cells (a population that tends to be underrated or even omitted from many studies) is the activation of antibody-producing B cells. Until 1990 antibody-mediated immunity was considered irrelevant in the host response to fungi, mainly due to the methods used thus far, such as passive transfer of immune serum, which could not be demonstrated to be effective. However, the technology of monoclonal antibody (mAbs) production has provided evidence for the role of specific antibodies to the benefit or detriment of the host [40]. The functions for combatting fungal pathogens include classical mechanisms such as phagocytosis and complement activation, but recent studies suggest additional mechanisms for antibodies, such as modulation of the inflammatory response, and inhibition of replication, germination, polysaccharide release, morphologic switch and biofilm formation, among others [40]. Likewise, protective antibodies against a wide variety of fungal pathogens, including C. neoformans, C. albicans, Histoplasma capsulatum, A. fumigatus, Paracoccidioides brasiliensis and Sporothrix schenckii, have also been described [41].
Different studies have demonstrated a strong and complex humoral response in immunocompetent and non-infected individuals to L. prolificans, mediated by both salivary IgA and serum IgG [42][43][44]. Specifically, salivary IgA recognizes almost exclusively the conidia of the fungus, while serum IgG recognizes both conidia and hyphae. This is consistent with the scenario of a fungal invasion of the respiratory tract, in which the host inhales the morphology used by the fungus for dispersal, the conidia, instead of hyphae [43]. Furthermore, taking into account the fact that some of the antigens identified in these studies are broadly conserved among fungal pathogens, this high reactivity observed for a healthy population may be related to a cross-reactivity process among these antigens and those of other common fungi, such as A. fumigatus [43][44].
These studies using sera and saliva from immunocompetent individuals are interesting; the fact that L. prolificans infections are almost exclusively suffered by immunocompromised individuals, although its environmental distribution is associated with humanized areas, seems to indicate that an immunocompetent immune system is able to control the fungus [43][44][45]. Some of the antibodies produced by these individuals may therefore be protective against a possible infection by L. prolificans, and the fungal antigens recognized could be therapeutic targets.
Some Scedosporium/Lomentospora antigens have been identified, including PRM and catalase, which could be interesting as diagnostic markers of these mycoses [46][47]. Among them, catalase A1 from S. boydii seems to be a candidate of special interest for serodiagnosis of infections caused by Scedosporium spp. in patients with CF [47].
Furthermore, during recent years several immunoproteomics-based studies have been published, enabling identification of a large number of Scedosporium/Lomentospora targets with potential medical applicability for the production of innovative treatment strategies, diagnosis or prevention of these fungal infections (Figure 4). Along these lines, Buldain et al. (2016) identified the cyclophilin and enolase as the most prevalent antigens of L. prolificans recognized by 85 and 80% of saliva from immunocompetent individuals, respectively [44]. These enzymes were also identified on fungal cell wall surface, on the immunomes of S. apiospermum and S. aurantiacum, and even exhibited cross-reactivity with A. fumigatus. These results show that the immune response might offer pan-fungal recognition of these conserved antigens, making them interesting candidates for therapeutic targets, since therapies directed against these enzymes could provide protection against different pathogenic fungi.
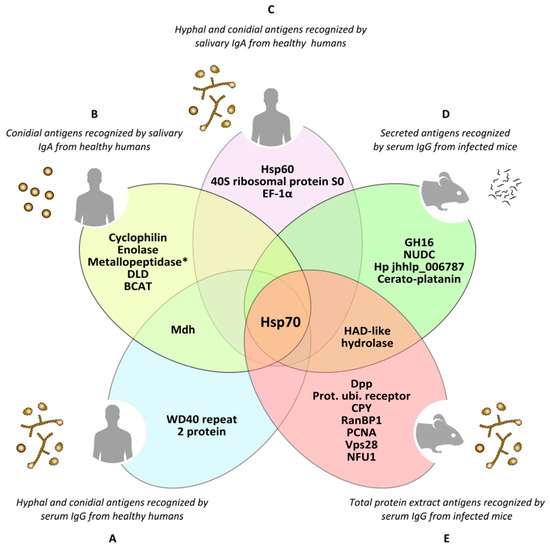
Figure 4. Antigens of Lomentospora prolificans identified in several immunoproteomics-based studies. Antigens detected as being reactive against at least 70% of human serum samples in both hyphal and conidia morphologies (A). Conidial antigens that reacted with at least 50% of human salivary samples (B). The most immunodominant antigens that react with human salivary samples, detected in both fungal morphologies (C). The most immunodominant antigens of secretome (D) and total protein extract (E) recognized by sera from infected mice. Mdh: malate dehydrogenase; Hsp70: heat shock protein 70; Metallopeptidase: Cys-Gly metallo-dipeptidase DUG1; DLD: dihydro-lipoyl dehydrogenase; BCAT: putative branched-chain-amino-acid aminotransferase TOXF; Hsp60: heat shock protein 60; EF-1α: translation elongation factor-1 α; GH16: glycosyl hydrolase family 16 protein; NUDC: nuclear movement protein nudC; Hp jhhlp_006787: protein with unknown function Hp jhhlp_006787; HAD-like hydrolase: halo-acid dehalogenase-like hydrolase; Dpp: dipeptidyl-peptidase; Prot. ubi. receptor: Proteasomal ubiquitin receptor; CPY: carboxypeptidase Y; RanBP1: RanBP1 domain-containing protein; PCNA: proliferating cell nuclear antigen; Vps28: vacuolar protein sorting-associated protein 28; NFU1: NFU1 iron-sulfur cluster scaffold-like protein. * This antigen has been added by repeating the peptides spectra analysis with a later version of the NCBI (National Center for Biotechnology Information) database.
In the case of immunocompetent individuals, Pellon et al. (2016) identified the proteins WD40 repeat 2, malate dehydrogenase (Mdh), and DHN1 in conidia of L. prolificans, and heat shock protein (Hsp) 70, Hsp90, ATP synthase β subunit, and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) in hyphae, as the most sero-prevalent antigens, detected by at least 90% of the human sera used [43]. Among them, Hsp70, Mdh and WD40 repeat 2 protein were identified in both morphologies. It is worth noting that ATP synthase α and β subunits, fructose-bisphosphate aldolase (Fba), Mdh, enolase and Hsp70 have also been identified as IgA-recognized antigens from saliva of healthy individuals [42][44]. Furthermore, relevant antigens such as Hsp70, enolase, and Hsp90 are present in the surface of fungal cell wall, increasing the interest of these sero-prevalent antigens as therapeutic and diagnostic targets due to their accessibility to the immune system [43].
More recently, Buldain et al. (2019) have demonstrated that sera from mice infected intravenously with L. prolificans, S. apiospermum and S. aurantiacum species, but not with A. fumigatus, showed a high cross-reactivity, which is most likely due to the phylogenetic proximity of Scedosporium/Lomentospora species [48]. This high cross-reactivity makes it impossible to find species-specific antigens inside Scedosporium/Lomentospora, but it may be very useful for differentiating this complex from Aspergillus. Among the antigens recognized by the sera of mice infected with these three species were: in L. prolificans, total protein extract, the proteasomal ubiquitin receptor, carboxypeptidase Y (CPY) and vacuolar protein sorting-associated protein 28 (Vps28); in the secretome, a protein of unknown function (Hp jhhlp_006787), a glycosyl hydrolase belonging to the GH16 family (GH16) and a cerato-platanin; and in both extracts, a halo-acid dehalogenase-like hydrolase (HAD-like hydrolase). Secreted proteins are of particular interest because they may be present in higher concentrations in body fluids and they share very little to no homology with human proteins [49][50]. In addition to diagnosis, these antigens may be of especial interest as therapeutic targets for studying alternative therapies for these infections, due to their null or low homology with their human homologues. Among all antigens identified in these studies (Figure 4), Buldain et al. (2019) stressed the importance of Scedosporium/Lomentospora Hsp70 for its reactivity, prevalence, location, and capacity to differentiate the infections generated by these species from A. fumigatus.
In the studies of humoral response carried out so far, a large number of targets of Scedosporium/Lomentospora with potential medical applicability have been identified, among which Hsp70, carbohydrate portion of PRM and catalase are of particular interest [51][46][42][43][44][45][47][48][52]. Nonetheless, so far only catalase A1 from S. boydii has been tested for clinical usefulness [47]. Although there is as yet no commercially available serological test for Scedosporium/Lomentospora, interest in the development of such detection systems is increasing. This is reflected in a recently-published article developing a useful ELISA test to detect Scedosporium/Lomentospora-infected patients and discriminate them from those infected with Aspergillus [28].
The mechanisms of the pathogenic fungi Scedosporium/Lomentospora for immune response evasion are largely unknown. However, in the studies performed so far, different fungal cell wall components involved in this process have been detected. Among them, the surface glycoprotein PRM of L. prolificans has an immunomodulatory effect by reducing the inflammatory response and increasing host non-protective response. Precisely, immunization with PRM in a murine model of invasive scedosporiosis generated a decrease in the secretion of proinflammatory cytokines and chemokines, and an increase in Treg cells and in IgG1 secretion, an immunoglobulin linked to a non-protective response. In this sense, PRM exacerbated the infection process, thereby facilitating colonization, virulence and dissemination of the fungus [38].
Moreover, species of Scedosporium/Lomentospora produce 1,8-dihydroxynaphthalene melanin (DHN-melanin), also found in A. fumigatus conidia [53]. This molecule is known to facilitate evasion of immune response by masking PAMPs, by interfering with phagolysosome formation and acidification and by inhibition of host cell apoptotic pathways [54]. However, Scedosporium/Lomentospora melanin function has been poorly studied so far. Specifically, Ghamrawi et al. (2014) studied the dynamic changes in S. boydii conidia depending on the age of cultures, demonstrating an increase in the amount of melanin over time and suggesting melanin polymerization. This polymerization progressively masks mannose-containing glycoconjugates, which are involved in immune recognition, perhaps allowing the fungus to escape from host immune defenses [53]. In another study, using mutants of L. prolificans lacking the dihydroxy-naphthalene (DHN)-melanin biosynthetic enzymes, the authors showed that melanin plays a protective role in the survival of the pathogen to oxidative killing and UV radiation [55]. From the few studies conducted, therefore, it may be concluded that melanin is an important virulence factor of Scedosporium/Lomentospora, since it masks fungal PAMPs by preventing fungal recognition and decreases oxidative killing by phagocytes [53][55].
On the other hand, it has been detected that Scedosporium/Lomentospora is capable of destructing host key proteins, which may eliminate host defenses and facilitate tissue invasion. S. apiospermum is thus able to efficiently degrade proteins such as the complement factors C3 and C1q in cerebrospinal fluid, presumably by proteolytic degradation [56]. Considering that complement proteins represent a major immune defense of the CNS and that the brain is one of the most affected organs following Scedosporium/Lomentospora dissemination [9][17], the ability to degrade complement factors might be implicated in the neurotropism of these fungi. An extracellular 33 kDa proteinase of S. apiospermum has also been detected which, like alkaline proteinase of A. fumigatus, is able to degrade human fibrinogen [57]. Furthermore, mycelia of S. boydii releases metallopeptidases capable of cleaving several proteinaceous compounds, including human hemoglobin, IgG extracellular matrix components (fibronectin and laminin) and sialylated proteins (mucin and fetuin) [58].
Enzymes implicated in oxygen species detoxification, catalase and Cu,Zn-superoxide dismutase (SOD), have also been also purified and characterized in Scedosporium spp. [59][60]. In particular, it was demonstrated that catalase A1 gene of S. boydii is overexpressed in response to oxidative stress and phagocytic cells, whereas the gene encoding SOD (SODC gene) is constitutively expressed [59].