Abstract
To gain an insight into fungal sooty mould communities on leaves of trees and shrubs in the tropics and in temperate regions, 47 biofilms of the Mata Atlântica rainforest relic and the Caatinga vegetation in the state of Sergipe, Northeast Brazil, and from Central European colline and alpine zones were compared. The four sampling sites clearly differed in composition of their epiphyllous fungal communities. The fungal OTUs from all sites belonged mainly to the Ascomycota, with Dothideomycetes being the dominant class. The core community group consisted of a few site-specific representatives in co-occurrence with the ubiquitous Mycosphaerella tassiana and Aureobasidium pullulans. Most species of the core community were dark pigmented and were accompanied by facultative unpigmented or lightly pigmented species. Among the cultivable fungal species, the proportion of melanised species was significantly more abundant in samples from the two European sites, which supports the theory of thermal melanism. The identity of the host plant had a stronger impact on fungal community composition than the presence of sap-feeding insects.
Similar content being viewed by others
Introduction
Surfaces of higher plants, particularly leaves and stems of trees and shrubs, are often colonized by dark pigmented fungi, which form hyphal mats. This type of colonization phenomenon is commonly referred to as the ‘sooty mould communities’ (Chomnunti et al. 2014; Kirk et al. 2010; Schoulties 1980), and can be observed in many habitats throughout the world. The absence of specific interbiotic cellular structures such as haustorial protrusions as present in truly phytopathogenic fungi (An et al. 2006; Dekhuijzen and Scheer 1969) indicates that these fungi have a predominantly or temporarily non-biotrophic life habit.
Sooty moulds are relatively common in tropical, subtropical, and warm temperate regions (Chomnunti et al. 2014). In the tropics, these (Faull et al. 2002; Olejnik et al. 1999) and other fungal associations exhibit considerable species diversity (Chomnunti et al. 2014; Dhami et al. 2013; Nelson 2008). The abundance of sooty moulds is highest during the rainy season (Batista and Ciferri 1963). Sooty moulds also occur in the cold and temperate climate zones of the European Alps, where they are formed by fungal communities with lower species numbers (Flessa and Rambold 2013).
A considerable number of sooty mould species show a clear preference for honeydew exudates of insects (Hughes 1976; Parbery and Brown 1986; Perez et al. 2009), such as aphids, whiteflies, soft scales, mealybugs, leafhoppers, and psyllids (Barr 1987; Chomnunti et al. 2014; Hamid and Jalaluddin 2006). Globally, more than 200 pigmented epiphyllous fungal species with members of the Antennulariellaceae, Capnodiaceae, Chaetothyriacae, Coccodiniaceae, Euantennariaceae, Metacapnodiaceae, and Trichomeriaceae have been recognised as belonging to this nutritional type. In their natural habitats, they may be accompanied by accessory, non-pigmented fungi (Chomnunti et al. 2014; Flessa et al. 2012; Flessa and Rambold 2013). Sooty mould fungi therefore form mixed associations with considerable proportions of pigmented species. In tropical environments, members of this community may further associate with representatives of the so-called black mildews, which belong to the Meliolaceae (Hansford 1961; Parbery and Brown 1986; Schoulties 1980) and are known as true plant parasites that develop intracellular haustoria in cells of host plants (Rodríguez and Piepenbring 2007). These obligate biotrophs interact closely with living plants to grow and reproduce, and are assumed to be host-specific (Hansford 1961; Hosagoudar 1996). Sooty mould communities are exposed to the entire airborne spore pool. Many spores can be deposited on leaves without being able to live there and an older community may therefore contain many spores that can be detected but do not contribute to the structure of the community.
To investigate the alpha diversity (Whittaker 1960) of sooty mould fungi in different continents and contrasting habitats, two sites in Brazil and two sites in Central Europe were analysed comparatively. The Brazilian sites are located in the state of Sergipe and are represented by two vegetation types. One is an open shrub vegetation with scattered trees (‘Caatinga’), which exhibit typical sooty mould infestations in combination with scale insects, and the other is a predominantly secondary Mata Atlântica relic rainforest. The two Central European sites represent open habitats in the colline (Germany) and alpine (Switzerland) zones.
The aim of the present study was to test the following hypotheses: (1) The diversity of sooty mould communities is higher in the tropical region than in the central European region and also varies among the individual geographical subregions. (2) The presence of sap-feeding insects (scale insects or aphids) supports the development of specifically composed communities. (3) The community composition is also triggered by other biotic site factors such as host plant species identity. (4) The structure of the communities at all sites is similar in that the predominant fungi are dark pigmented and saprotrophic.
Material and methods
In order to investigate the differences in α-diversity of the sooty mould community at the study sites and in the two regions Central Europe and Brazil, community barcoding (CB) was performed. Since HTS does not distinguish between living and dead fungi and does not allow for a direct investigation of pigmentation, the results were complemented by a cultivation-based single-species barcoding (SB) approach. In addition, the cultivation-based approach can exclude strict biotrophs.
Sampling and conceptual design of the study
A total of 47 samples were examined with community barcoding (CB) and 33 samples with single-species barcoding (SB). In order to compare samples from ecologically distinct habitats, two sites in the neotropics (North-eastern Brazil) and two sites in the temperate zone (Central Europe) were included in this study. The two sites in the Brazilian state of Sergipe are located at lower altitudes but have two different vegetation types: Site 1 (16 samples for CB, 10 samples for SB): North-eastern Brazil Caatinga (BC): Brazil. Sergipe, Areia Branca, Park dos Falcões, 40 km NW of Aracaju, 7 km SE of Itabaiana, c. 7 km in north-easterly direction from Areia Branca on the road BR235 from Aracaju to Itabaiana, beside rough road at entrance of the park. Open habitat with scattered trees or shrubs. Fungi from sap-feeding insects were also studied and are labelled BC* (4 samples for CB). Site 2 (16 samples for CB, 10 samples for SB): Northeast Brazilian Tropical Rainforest (BR): Brazil. Sergipe, Areia Branca, Park National Serra de Itabaiana, 36 km NW of Aracaju, 10 km SE of Itabaiana, 3 km northeast of Areia Branca on road BR235 from Aracaju to Itabaiana, beside rough road through the park area. Relic forest (Mata Atlântica rainforest) with tall trees and shrubs. Site 2 is located at a linear distance of 4 km from site 1. The two sites in Central Europe mainly differ in altitude and corresponding vegetation type: Site 3 (6 samples for CB, 8 samples for SB): Central Europe alpine zone (EA): Switzerland. Canton Bern, Grimsel Valley, on Rhododendron ferrugineum shrubs (none with sap-feeding insects). For a further characterisation of the sampling sites, see Flessa and Rambold (2013). Site 4 (5 samples for CB, 5 samples for SB): Europe colline zone (EC): Germany. Upper Franconia, Bayreuth, campus area and Ecological-Botanical Garden of the University of Bayreuth. For more details on the sampling sites, see Flessa et al. (2012).
Host plant sampling data are included in Supplementary Data S1. Host plants of sites BC and BR were assigned to their respective host plant identity groups based on DNA sequence analysis data. The protocol is referred in the “DNA isolation” and “PCR and sequencing” sections. The species identities of the host plants Rhododendron ferrugineum L. and Salix spp. from sites EA and EC were confirmed or revised based on morphological traits. The nomenclatural and classification concepts for the host plants follow the Catalogue of Life Annual Checklist 2019 (Roskov et al. 2019).
Associated sap-feeding insect identification: The presence or absence of the insects was recorded during the sampling period. If the insects were still on the plant during collection, they were examined microscopically and determined by morphological traits, and if amplicons of the mitochondrial cytochrome oxidase I (COI) gene were obtainable, individuals were assigned to their respective taxon. The protocol is referred to in the “DNA isolation” and “PCR and sequencing” sections.
The average annual temperature in Brazil (Itabaiana) is 25.4 °C, which differs from the European colline site with 7.8 °C and the European alpine site with 1.9 °C (climatic data resources: https://de.climate-data.org/suedamerika/brasilien/sergipe/itabaiana-42970/;http://www.bayceer.uni-bayreuth.de/meteo/de/klima/gru/html.php?id_obj=139937;https://tirolatlas.uibk.ac.at/graphics/lieth/diag.py/chart?id=1370932).
Hypotheses 1–3 were tested primarily using the CB approach, while the SB approach was used for testing the supplementary hypothesis. Diversity indices, NMDS and PERMANOVA, were used to investigate the influence of the four sites, the two continents, the occurrence of sap-feeding insects, and the host plant family and order on the composition and diversity of the sooty mould communities. At the BC site, the sooty mould community on leaves (BC) was additionally compared with the insect-associated fungi (BC*). Overlaps and differences in the species spectrum are shown using Venn diagrams; the dominant species spectrum is shown using bar plots and a heatmap. For the most common ascomycetes from both approaches, the nutrition type was determined with FUNGuild, compared with literature, and the result visualized with iTOL Tree. To determine the proportion of dark pigmented saprotrophs, the pigmentation was examined for the most common fungi of the SB approach.
Sampling dates and sample treatment
For culture-independent community barcoding, material was collected at the four sites in October 2013. Fragments of mycelia were cut from infected leaf surfaces in the field with tweezers and scalpel, placed in 1-ml screw cap tubes, and brought to the laboratory. The tubes were kept at a temperature of ± 0 °C until DNA isolation which took place within a few days.
For species barcoding, biofilms of sooty mould communities from site EC were sampled from surfaces of young twigs of five species of Salix species in May 2006, as the host plants have annual leaves (Flessa et al. 2012). Biofilms of EA sooty mould communities were collected from visibly infested leaves in August 2007 (Flessa and Rambold 2013). Host plants with sooty mould communities on leaves at the BC and BR sites were sampled in October 2009. Branches of the plants were cut off, individually packed in vouchers, and transported to the laboratory. Cultivation took place immediately afterwards within a few days.
Cultivation and pigmentation test
Mycelial fragments were washed in sterile tap water, and transferred to Petri dishes containing malt-yeast medium (MYA) with 4 g glucose, 10 g malt extract, 4 g yeast extract, 0.1% tetracycline, and 12 g agar per litre. Cultures from EC, BR, and BC were incubated at room temperature and examined daily for mycelial growth. As the fungi of EA samples grew at temperatures of 15 °C or below, the incubation temperature for the fungal strains was set to 15 °C (Flessa and Rambold 2013). Outgrowing hyphae were transferred to new plates until pure cultures were obtained. Pure cultures were sorted according to their phenotypic traits (i.e. presence and type of aerial mycelia, growth form, and pigmentation type). From all isolated strains, at least one representative of each morphotype was selected for sequencing. One fungal strain per operational taxonomic unit (OTU) with more than two members and an assortment of singletons was deposited in the Jena Microbial Resource Collection (JMRC Jena). The accession numbers of the deposited strains are in Supplementary Data S2.
Pigmentation tests: Fungi were classified by their hyphal pigmentation using cultures grown on malt-yeast agar medium (MYA). Fungal cultures from EA were incubated at 15 °C and cultures from EC, BR, and BC at room temperature and examined after 1 month for macroscopically recognisable pigmentation. The pictures were taken with a Nikon D2x and lens Micro-Nikkor 60 mm F/2.8 D, with a distance of 36.5 cm to the object. The lighting was performed with two neon lamps (L18 W/19 Daylight 5000 de Luxe), which were attached to a frame with 80° inclination and provided constant lighting of the images. A colour chart (RAL 7005-HR mousy; German Institute for Quality Assurance and Certification e. V.) was used as the standard background. Settings: lens aperture of 1/16, sensitivity ISO 100, balanced preset d-0, colour mode III (Adobe RGB), and shutter speed of 0.5 s (Pietrowski et al. 2010). The CMYK values of the digital images for testing the degree of pigmentation were classified as ‘dark’: C: 39–73%; M: 50–75%; Y: 51–94%; K: 21–83%. CMYK values for pigmentation classified as ‘light’ or ‘absent’: C: 28–49%; M: 32–48%; Y: 33–61%; K: 0–10% according to Flessa and Rambold (2013).
DNA isolation
DNA from fungi, host plants (from BC and BR), and insects was isolated using the Charge Switch® gDNA Plant Kit (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA). Cell disruption was performed using a Fast Prep FP120 (Bio101, Thermo Fisher Scientific Inc., Waltham, MA, USA) with a mixture of glass beads, consisting of 0.3 g of beads with a size of 0.1 mm, 0.5 g with a size 0.5 mm, and three glass beads with a size of 2.8 mm (Peqlab) at a speed of 6.0 m s−1 for 2 × 40 s.
PCR and sequencing
For community barcoding, the amplification steps during the preparation of the internal transcribed spacer (ITS) library preparation for the Illumina sequencing included a two-step PCR using the ITS1F/ITS4 forward and reverse primers (Gardes and Bruns 1993; White et al. 1990) combined with specific tag sequence combinations (TAG) and the Illumina sequencing primer to amplify the ITS1 and ITS2 regions of the rDNA gene (Supplementary Data S3a). The first PCR step was performed using 30 cycles of denaturation at 94 °C for 27 s, annealing at 57 °C for 1 s, and extension at 72 °C for 90 s. The second PCR step was performed after a purification step using Exonuclease I, with an activity of 2 U/ml (New England Biolabs), and Shrimp Alkaline Phosphatase, with an activity of 0.1 U/ml (New England Biolabs), and included 5 cycles of denaturation at 94 °C for 27 s, annealing at 53 °C for 1 s, and extension at 72 °C for 90 s, using the Illumina sequencing primer, combined with a second tag combination (INDEX) and the Illumina P5 or P7 sequencing adapters, respectively (Supplementary Data S3b). All PCR steps were performed using a Primus 96 plus thermal cycler (MWG Biotech, Ebersberg, Germany). Final amplicons were purified, quantified by gel staining with ethidium bromide in a 0.8% agarose gel and 100% UV light under a GelDoc Station (MWG Biotech), and finally pooled over several steps to minimize variations in total amplicon concentrations between samples. Paired-end sequencing was performed using an Illumina MiSeq® sequencer at the Ludwig Maximilian University of Munich Biocenter (Illumina Inc., 2018). For the list of used primers, see Supplementary Data S3.
For species barcoding, host plants, and insects, PCR was performed using MWG Biotech Primus 96 plus thermocycler. Using the Charge Switch® PCR Clean-Up Kit (Invitrogen), the obtained double-stranded sequences of the fungal ITS rRNA gene were purified and further processed according to Triebel et al. (2005) using primers ITS4 (White et al. 1990) and ITS1F-Z (5′-CTWGGYYATTTAGAGGAAGTAA; modified after Gardes and Bruns 1993). Amplicons of the host plant RNA polymerase C1 gene (rpoC1) fragment were obtained from 5 μl purified PCR products (20–80 ng/μl) with 5 μl of primers (5 pmol/μl) by GATC Biotech, by Sanger sequencing using primers rpoC1-4r (5′-CCA TAA GCA TAT CTT GAG TTG G) and rpoC1-2F (5′-GGC AAA GAG GGA AGA TTT CG) (Hollingsworth et al. 2009; Saddhe et al. 2017). Amplicons of the insect COI fragment (Folmer et al. 1994) were obtained from 5 μl purified PCR products (20–80 ng/μl) with 5 μl of primers (5 pmol/μl) by GATC Biotech using Nancy and Ron primers (Simon et al. 1994). DNA reference sequences were obtained by Sanger sequencing; consensus sequences were deposited at NCBI GenBank under accession numbers GU942834–GU942923 (fungal ITS), KM259875–KM259899 (host plant rpoC1 from BC and BR), and MN604915-MN604921 (insect COI).
Bioinformatics
For community barcoding, the reads obtained from Illumina sequencing were quality-checked using FastQC (Andrews 2010) and demultiplexed to sample-level by the variable TAG sequences, with all reads containing ambiguous base-calls removed using the pipeline Quantitative Insights Into Microbial Ecology ver. 1 (QIIME1) (Caporaso et al. 2010). Demultiplexed reads were then imported into QIIME ver. 2 (QIIME2) (Bolyen et al. 2019) and trimmed using the ‘cutadapt’ plugin (Martin 2011), thereby eliminating any remaining Illumina barcodes and primer sequences. To obtain amplicon sequence variants (ASVs) (Callahan et al. 2017), the DADA2 denoiser was applied as a QIIME2 plugin for dereplication, removal of all detected chimeric sequences, and quality-filtering, at a maximum expected error rate of 2 (Callahan et al. 2016). Those ASVs were further processed into OTUs (de novo) at a 97% similarity by passing their representative sequences to the VSEARCH tool, embedded in the QIIME2 pipeline (Ezeokoli et al. 2020; Rognes et al. 2016). OTU clustering was followed by an additional chimeric filtering step, applying the UCHIME implementation using the VSEARCH tool. Contextual data concerning the project and the sampling were uploaded to the NCBI databases BioProject and BioSamples. Illumina sequencing results were deposited at the NCBI ‘Sequence Read Archive’ (SRA) database; the uploaded demultiplexed sequences are available via the respective link from the BioProject entry with the accession number PRJNA699866. For species barcoding, fungal isolates were grouped for further statistical analysis based on their ITS rRNA sequence similarities as described by Peršoh et al. (2010). Pairwise similarities among length-adjusted sequences were calculated by using the BLAST application ‘blastall’ (v. 2.2.18). The resulting tabular output dataset was transformed with the simMatrix function, using the RFLPtools package (Flessa et al. 2010) of R (R, Core Team 2020). Based on this dataset, cluster analysis was performed in R with the function ‘hclust’ using the clustering method ‘average linkage’, and clusters with minimal similarities of 97% (Flessa et al. 2012) were grouped into species barcoding operational taxonomic units. The occurrence of fungal groups was noted for the samples of each host plant, yielding the present/absent matrix for further statistical analysis.
Taxonomy assignment of fungal OTUs and strains
Using the UNITE database as a reference, a naïve Bayesian classifier was trained on the UNITE v.8.0 dynamic dataset (Nilsson et al. 2019) and used to assign the taxonomy to fungal OTUs via the QIIME2 pipeline applying the command feature-classifier classify-sklearn with a confidence threshold of 70% required for taxonomy assignment.
Statistical data analysis
The frequency table, its taxonomy, and metadata (in which the geographic site and occurrence of leaf-sucking insects as well as the phylogenetic affiliation of the host plant taxon were encoded as grouping factors) of community and species barcoding approaches were converted to CSV format, imported in R v.3.6.2 (R Core Team 2020), and subsequently merged to an S4 object using ‘phyloseq’ v1.30.0 (McMurdie and Holmes 2013). Unless otherwise stated, all evaluations and analyses were performed with R. Plots of rarefaction curves were built with the ‘ggrare’ function in the ‘ranacapa’ package v. 0.1.0 (Kandlikar 2019). A dissimilarity matrix on relative abundance was calculated using the dissimilarity method most appropriate for the data obtained with the ‘rankindex’ function in the ‘vegan’ v.2.5–6 package (Oksanen et al. 2019), i.e. Manhattan dissimilarities. Variation of fungal communities between samples for community barcoding was visualized by non-metric multidimensional scaling (NMDS) in the ‘vegan’ package using the ‘metaMDS’ function and plotted with package ‘ggplot2’ v.3.2.1 (Wickham 2016). For the species barcoding dataset, Jaccard similarity was calculated and NMDS was visualized in Primer 6 (v. 6.1.11, Primer-E Ltd., UK). Permutational multivariate analysis of variance (PERMANOVA) was performed on both approaches with the ‘adonis’ function in the ‘vegan’ package to compare fungal communities based on the factor sampling site, continent, sap-feeding insect occurrence, host plant family, and order. To perform pairwise comparisons between individual factors, pairwise PERMANOVA was conducted using the package ‘pairwiseAdonis’ v.0.0.3 (Martinez 2019) for significance testing (p < 0.05) applying Benjamini-Hochberg false discovery rate adjustments on all p values to protect against false positives (Benjamini and Hochberg 1995; Ricks and Koide 2019). It was assumed that the p value indicated significant differences if it was less than 0.05. The ‘adonis’ function calculates an R value that indicates the extent to which the factors analysed explain the groupings. R values were interpreted as follows: > 0.75 indicating different groups, R > 0.5 overlapping different ones, and R < 0.25 indicating slightly separate groups.
Venn diagrams were drawn using ‘VennDiagram’ v.1.6.0 (Chen 2018). Alpha diversity indices, i.e. Chao1 richness estimate, Shannon-Wiener diversity index, Simpson’s dominance, and Pielou’s evenness, were calculated with the ‘alpha’ function in the package ‘microbiome’ v1.8.0 (Lathi and Shetty 2017) and plotted with ‘ggplot2’. Bar plots of phylum, class, and order were drawn using the ‘tax_glom’ function in the package ‘phyloseq’ and plotted with package ‘ggplot2’. Heatmaps were plotted from all OTUs, which were present in at least 10% of all samples using the package ‘pheatmap’ v.1.0.12 (Kolde 2019) and ‘RColorBrewer’ v.1.1–2 (Neuwirth 2014). All analyses described in this section were performed on all OTUs without any further selection; heatmaps were based on those OTUs, which were present in at least 10% of all samples.
Fungal guild nutrition type of the predominant Ascomycota
From the OTUs belonging to the Ascomycota, those that were present in at least 3 samples and had a taxonomic assignment at genus or species level and a guild assignment at species, genus, or family level were selected using the FUNGuild database (Nguyen et al. 2016). If there was no entry in the database, the nutrition type as recorded in the Dictionary of Fungi was used (Kirk et al. 2010). If there was no entry in either source, OTUs from community barcoding were classified as ‘n.a.’ (not assignable). For strains from species barcoding, mixotrophic or eventually saprotrophic nutrition type was assigned due to their cultivability. In addition to the rough classifications into saprotrophic, mixotrophic, and biotrophic nutrition types, the following life habit subtypes were applied: P: plant-associated (including litter saprotrophic and wood saprotrophic); A: animal-associated; S: saprotrophic; and O: ‘other’ (i.e. non-specific, fungus-associated, lichen-associated, dung saprotrophic); U: non-specific saprotrophic. The representative sequences were aligned using Mega7 software (Kumar et al. 2016) and a maximum likelihood phylogenetic tree (Egidi et al. 2019) was built using a Kimura 2-parameter model. For the visualization, the tree was uploaded to iTOL V5.5 (Letunic and Bork 2019) and supplemented with the nutrition type information and taxonomic classification.
Results
Dataset descriptions
Sooty mould symptoms were formed by oligo-species communities with an average of 11.2 species per biofilm sample from culture-based species barcoding and of 26.7 OTUs per sample from culture-independent community barcoding. Single-species communities are almost non-existent. In both approaches, > 60% of all OTUs could be assigned at order level or higher, and about one-third at species level. The proportion of OTUs that could not be assigned at all is below 26%. The rarefaction curves generated for each sample showed saturated curves for the community barcoding approach (Supplementary Data S4.1a). For a summary of the number of OTUs and their occurrence as well as a list of taxa detected by one or both approaches, see Supplementary Data S5.
Impact of the factors site and continent (community barcoding)
Differences in alpha diversity indices: The average expected number of OTUs per sample is highest in North-eastern Brazil Caatinga (BC) and the Europe Alpine (EA) zone, and lowest in scale insects from BC (BC*), whereas the difference in the number of species between the samples of BC* is the lowest, and the greatest in the Central European colline (EC) zone (Fig. 1a). The expected number of OTUs per sample is similar in both continents. The Shannon-Wiener index shows differences in the diversity of OTUs at all sites, with the two European sites (EA and EC) clearly differing from the Brazilian ones (Fig. 1a). Less OTUs dominate in Brazilian samples than in European ones, as the two European sites have considerably lower dominance values than the sites in South America, with EC samples differing more markedly from the others. European colline (EC) and Brazil rainforest (BR) show a higher degree of evenness in the composition of their fungal communities than Brazil Caatinga (BC) and Europe Alpine (EA) (Fig. 1a). While 5 OTUs are shared by all sites, another 6 occur at three sites each. BC and BR have the highest number of exclusively shared OTUs among all pairs, with 30 OTUs. However, most OTUs can only be found in samples of one site (from 75 in EA to 184 in EC). Thirty-seven OTUs occur in samples on both continents (Supplementary Data S4.2). The NMDS on fungal community similarity showed that BR samples are the most widely dispersed (Fig. 2), the two European sampling sites are clearly separated, and EC is close to BC and shows overlap with BR, but this overlap is due to a single sample of BR with few reads. At a threshold of 95%, all sampling sites overlap. PERMANOVA yielded significant results (p = 0.001) for differences in community composition between sites and continents (Table 1). However, as the r2 value of the factor ‘continent’ (r2 = 0.101) is very low, it explains the differences in community composition less well than the factor ‘site’. The pairwise PERMANOVA between the groups of factor ‘site’ showed significant results between all site pairs (Table 1) for both approaches.
Alpha diversity estimates of fungal communities a among the sites North-eastern Brazil Caatinga leaves (BC) and sap-feeding insects (BC*), North-eastern Brazil Tropical Rainforest (BR), Europe alpine zone (EA), and Central Europe colline zone (EC). b Continents Europe (EU) and South America (SA). c Association with sap-feeding insects (SFI+ = sap-feeding insect presence, SFI− = no sap-feeding insect association)
Non-metric multidimensional scaling (NMDS) ordination plot showing the compositional differences of the sooty mould communities from the different sampling sites North-eastern Brazil Caatinga leaves (BC) and sap-feeding insects (BC*), North-eastern Brazil Tropical Rainforest (BR), Europe alpine zone (EA), and Central Europe colline zone (EC). Confidence interval of ellipses = 95%, Manhattan dissimilarities, stress = 0.158
Impact of sap-feeding insects (community barcoding)
Based on the comparison of the community composition on what is the main nutritional source provided by the habitat, three types of sources could be distinguished: exudates by secretion from leaf glands, substances by leaching from leaves, and exudates from leaf-sucking insects. While leaching occurs to some extent on every host plant, glandular secretion was confirmed only for the EA host plant. Traces or remains of sap-feeding insects were confirmed in the field for leaves of host plants from BC. In EC, traces or remains of sap-feeding insects (Aphididae) were observed on all sampled trees in the field. No leaf-sucking insects or traces thereof were found at BR and EA sites. The grouping factor ‘sap-feeding insect association’ leads to a similar number of expected OTUs per sample, similar diversity, and only slightly different dominance and evenness between the two groups (Fig. 1c). Of all OTUs, 58 occur in both groups (Supplementary Data S4.2). Differences in community composition under the factor ‘sap-feeding insect association’ showed a significant impact (p = 0.001) in the PERMANOVA. However, as the r2 value of this factor (r2 = 0.092) was very low, it explains the differences in community composition less well than other factors. For the BC site, samples of sooty moulds from leaves and fungal OTUs from sap-feeding insects on infected plants (labelled as BC*) were compared. The communities found on and in sap-feeding insects showed complete overlap with those of leaf samples and proximity to the EC samples in the NMDS analysis. The pairwise PERMANOVA between groups of the factor ‘site’ showed significant results between all pairs except between BC and BC* and BC* and EC (Table 1), which is also consistent with the NMDS analysis.
Host plant dependence (community barcoding)
PERMANOVA revealed significant differences (p = 0.001) in community composition between host plant families and orders (Table 1). Significant differences were found in the pairwise comparisons between 9 host plant orders. The corresponding table of results is given in Supplementary Data S6, and the pairwise comparisons of host plant families in Supplementary Data S7. Sooty mould communities could not be found on all plants of a site, and the host plant families and orders were strongly linked to the sites, i.e. Asterales and Dilleniales occurred only at BC, Ericales only at EA, and Gentianales and Sapindales only at BR. This factor should therefore be viewed with some caution. The fungal community compositions differ significantly (r2 = 0.304, p = 0.003) when ‘host plant order’ and ‘site’ are tested as a combined factor, but there are no significant results in the pairwise tests (data not shown). The community compositions are also significantly separated based on the paired factor ‘host plant order’ or ‘host plant family’ with ‘sap-feeding insect association’ (Table 1). Although the global PERMANOVA gives a high r2 value for host plant family and sap-feeding insect, none of the pairwise PERMANOVA tests is significant (data not shown). Pairwise tests of host plant order and sap-feeding insect between 9 groups were significant. The corresponding table of results is given as Supplementary Data S6.
Taxonomy (community barcoding)
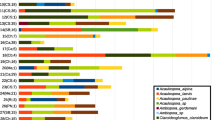
While OTUs were used anonymously for all previous analyses, the analyses in the following section are based on taxonomic assignments. Ascomycota and Basidiomycota were found at all sites, with Ascomycota predominating (Fig. 3a). Within the Ascomycota, Dothideomycetes were the dominant class at all sites, followed by Eurotiomycetes and Sordariomycetes, which were present at all four sites (Fig. 3b). Within the Dothideomycetes, the Capnodiales and Pleosporales dominated and were also present at all sites. Capnodiales are the dominant order, with a proportion of over 50% in BC incl. BC* and EA, over 25% in EC, and over 10% in BR, considering the high proportion of unassignable ones in BR. Both European sites are characterised by a higher relative abundance of Eurotiomycetes. Among the Sordariomycetes, the Hypocreales and Xylariales were the dominant order, but were mainly found in samples from BC and less frequent in BR. The Xylariales were absent in the group ‘sap-feeding insects’ (BC*) and were found in EA and EC in only one sample each. Within Basidiomycota, Tremellomycetes were predominant and were found at all sites (Fig. 3c) but their relative abundance is higher at BC and EC sites than at the other sites. BR samples showed a higher relative abundance of unassignable OTUs (NA). The steadiness of OTUs that were present in at least 10% of all samples is summarized in Fig. 4. The Brazilian sites differ from the two European sites in community structure. However, the two European sites are less similar than the Brazilian ones. Mycosphaerella tassiana, Aureobasidium pullulans, and Vishniacozyma sp. Pleosporales sp. occurred in BC, BR, EA, and EC. Capnodiaceae sp. and Capnodium sp. occurred only at the Brazilian sites, and Capnodiales sp. 3 and Sarcinomyces sp. only in EA samples. Capnodiaceae sp. and Capnodium sp. tend to co-occur. This also applies to Sarocladium strictum and Alternaria sp. together with Erythrobasidium hasegawaianum and Capnodiales sp. with Sarcinomyces sp.
Relative abundance of a phyla (Ascomycota and Basidiomycota), b class (Ascomycota: 11–19, Basidiomycota: 20–29), and c order (11: Arthoniomycetes; 12–17: Dothideomycetes; 18–20: Eurotiomycetes; 21–25: Lecanoromycetes; 26–29: Leotiomycetes; 30: Orbiliomycetes; 31: Saccharomycetes; 32–39: Sordariomycetes; 40: Taphrinomycetes; 41–43: Agaricomycetes; 44: Agaricostilbomycetes; 45–46: Cystobasidiomycetes; 47–48: Exobasidiomycetes; 49: Malasseziomycetes; 50–52: Microbotryomycetes; 53: Pucciniomycetes; 54–58: Tremellomycetes; 59: Ustilaginomycetes; 60: Wallemiomycetes) in North-eastern Brazil Caatinga leaves (BC) and sap-feeding insects (BC*), North-eastern Brazil Tropical Rainforest (BR), Europe alpine zone (EA), and Central Europe colline zone (EC) samples
Heatmap of the most common taxa based on their consistency in North-eastern Brazil Caatinga leaves (BC) and sap-feeding insects (BC*), North-eastern Brazil Tropical Rainforest (BR), Europe alpine zone (EA), and Central Europe colline zone (EC) samples. The colour scale illustrates the dominance (%) within the sites. The dendrogram on top illustrates the similarity of the sites; the one on the left represents the similarity in the pattern of occurrence of the individual fungal OTUs
In order to classify the trophic mode within the Ascomycota via FUNGuild, those OTUs were selected that were present in at least three samples and that had a taxonomic assignment to species or genus level. The criteria applied to 29 OTUs, representing 4.27% of all OTUs in this dataset and 37.17% of all reads, which were combined in an iTOL tree (Fig. 5). Thirteen of these OTUs have a biotrophic trophic mode, 9 are classified as saprotrophic, and another 7 are declared mixotrophic according to FUNGuild. Three-quarters of OTUs are plant-associated, 5 of them are also animal-associated. All those belonging to the saprotrophic type were classified as unspecific. None of the OTUs was exclusively animal-associated.
Most abundant OTUs (occur in at least 3 samples) among the Ascomycota with taxonomic assignment on species or genus level and with FUNGuild classification on species, genus, or family level. Tree was generated with maximum likelihood method by Mega7 and visualized with iTOL (interactive Tree of Life). P, plant-associated (including litter saprotrophic and wood saprotrophic); A, animal-associated; S, saprotrophic; O, ‘other’ (i.e. unspecific, fungus-associated, lichen-associated, dung saprotrophic)
Single-species community barcoding
The complementary analyses of cultivable fungi showed similar results to the community barcoding in terms of the influence of the factors ‘site’, ‘continent’, ‘sap-feeding insect association’, and ‘host plant family and order’. All factors examined were significant. Again, the r2 values for the grouping factor ‘continent’ (r2 = 0.163) and sap-feeding insect presence (r2 = 0.09) were lower than those of the other factors (Supplementary Data S8.2a). In contrast to community barcoding, BC and BR samples overlap completely in the NMDS, while EA and EC are separate and do not overlap with Brazil samples (Supplementary Data S8.1, 8.3c–5c). Pairwise PERMANOVA between site factor groups showed significant results between all pairs except for the two Brazilian ones (Supplementary Data S8.2b), which is consistent with NMDS analysis. The cultivable part of the European communities is represented by a much lower number of species per sample than of the South American ones. One species was found in all four sites, most (37) are shared by BC and BR (Supplementary Data Fig. S8.4b). Ascomycota and Basidiomycota were found at all sites, with Ascomycota predominating (Supplementary Data Fig. S8.6a). The ratio Ascomycota to Basidiomycota is similar to that of community barcoding. The order Capnodiales within the Dothideomycetes is also the group with the highest relative abundance (Supplementary Data Fig. S8.6b–c). Likewise, also the Sordariomycetes show a higher abundance in BC and BR. Among the orders, the relative proportion of Pleosporales is considerably higher at all sites. This also applies to Xylariales in the BR and BC samples (Supplementary Data Fig. S8.6b–c). In contrast to community barcoding, only Aureobasidium pullulans occurs at all four sites; Mycosphaerella tassiana and Didymella exigua occur in a large number of samples from BC, BR, and EC, and Antennariella placitae mainly in BC samples and to a lesser extent in BR and EA samples. In accordance with the results obtained with community barcoding, Capnodiales sp. was found exclusively at the EA site and there in 85% of the samples (Supplementary Data Fig. S8.7). Trophic modes according to FUNGuild for the most common Ascomycota are given in Supplementary Data S8.8.
Pigmentation of sooty mould fungi
The degree of pigmentation was determined by taking into account the steadiness of the species. The proportion of detections of dark pigmented fungal species was 46.2% across all fungal species. Of those present in at least one-third of all samples, 60% were dark pigmented, and of those present in at least half of all samples, 100% were dark pigmented. This analysis shows that the more common fungi at each site were dark pigmented. At all four sites, the proportion of dark pigmented species varied widely, with the highest value in EC (60%) and lower values in BC (47.5%) and EA (46.3%), and lowest in BR (43.8%). Among the predominant which were present in at least one-third of all samples, the ratio shifts towards the dark pigmented ones with the highest values in EA (100%) and EC (67%) and slightly lower proportion in BC (59%) and lowest value in BR (48%). A trend for a correlation between nutrition type and pigmentation could not be found (Fig. 6) neither generally for all fungi nor for those found in at least one-third of the samples per site. The dark pigmented fungi of this study could all be assigned to Ascomycota, while all Basidiomycota were light pigmented (Supplementary Data Fig. S8.9). Dothideomycetes were the largest class among the dark pigmented and Sordariomycetes represented the largest group among the light pigmented ones.
Proportion of pigmented species and link between pigmentation and nutrition type. a Proportion of dominant pigmented species (present in at least 1/3 of all samples at this site) taking into account their steadiness. North-eastern Brazil Caatinga leaves (BC) and sap-feeding insects (BC*), North-eastern Brazil Tropical Rainforest (BR), Europe alpine zone (EA), and Central Europe colline zone (EC). b All species regardless of their dominance from species barcoding are grouped by pigmentation and nutrition types based on the relative amounts of samples they were found in
Discussion
Applied methods
This study used a cultivation-based species barcoding approach (SB) and a community barcoding approach (CB). With regard to the significant impact of the factors site, continent, sap-feeding insect presence, and host plant family and order on fungal community composition, the results for both approaches are similar. For community barcoding, the ITS1 region was used as marker. In recent studies, the use of the two ITS regions has been quite inconsistent. While some used ITS2 as the universal fungal barcode (Bao et al. 2019; Beng and Corlett 2019; Egidi et al. 2019; Epstein et al. 2019; Li et al. 2019; Lynikiene et al. 2020; Morales-Rodriguez et al. 2019; Nerva et al. 2019; Qian et al. 2020; Ricks and Koide 2019; Saravesi et al. 2019; Wilkinson et al. 2019; Würth et al. 2019; Yao et al. 2019; Zhang et al. 2019), others strongly recommended the use of ITS1 (Mbareche et al. 2020), as it had already been implemented in many studies (Beule et al. 2019; Checinska Sielaff et al. 2019; Del Frari et al. 2019; Schiro et al. 2019). Few studies are based on both ITS regions (Janakiev et al. 2019). The use of operational taxonomic units (OTUs) versus the recent use of amplicon sequence variants (ASVs) is also inconsistent. Although there are recommendations to use only ASVs (Callahan et al. 2017), resulting in a reduction in the number of dominant species (Egidi et al. 2019), many of the current studies are still based on OTUs (Bao et al. 2019; Beng and Corlett 2019; Beule et al. 2019; Checinska Sielaff et al. 2019; Epstein et al. 2019; Janakiev et al. 2019; Lynikiene et al. 2020; Morales-Rodriguez et al. 2019; Nerva et al. 2019; Pan et al. 2019; Qian et al. 2020; Ricks and Koide 2019; Saravesi et al. 2019; Wilkinson et al. 2019; Würth et al. 2019; Yao et al. 2019; Zhang et al. 2019), but others rely on ASVs (Del Frari et al. 2019; Egidi et al. 2019; Li et al. 2019), or OTUs generated from ASVs (Ezeokoli et al. 2020), and even OTUs from cultivation-based Sanger sequencing are still in use (Janowsky et al. 2019). With the availability of the VSEARCH tool within the QIIME2 pipeline, allowing the easy aggregation of OTUs from ASVs, the Illumina sequencing datasets in this study were analysed based on ASVs, obtained from the DADA2 plugin from QIIME2, with subsequent grouping into OTUs, based on a sequence similarity threshold.
By including a culture-based method, it was possible to include the degree of pigmentation of the mycelia as an analytical factor in the design of this study, to detect the fungi that are mixotrophic and to exclude those which are exclusively biotrophic. Those aspects cannot be addressed by the exclusive use of a culture-independent approach. Also, the fungi found with the different approaches may differ (Dissanayake et al. 2018). To put the significance of the pigmentation tests into perspective, it must be taken into account that pH, carbon, and nitrogen source have a major influence on pigment production in fungal strains (Lathadevi et al. 2014). Therefore, pigmentation may differ in culture and in situ. It is well documented that the degree of pigmentation of fungal cultures depends on the medium (Kowalski et al. 2016; Palacio-Barrera et al. 2019; Blechert et al. 2019). In a previous study of sooty mould fungi associated with R. ferrugineum, fewer fungi were dark pigmented on a MYA medium than on other media (Flessa and Rambold 2013). However, any fungus that was dark pigmented on MYA was also dark pigmented on the other media. It can therefore be assumed that the proportion of dark pigmented fungi of sooty mould communities in situ is possibly underestimated.
Host plants of tropical parasitic black mildews (Meliolaceae) in Brazil are known to belong to Anacardiaceae, Asteraceae, Euphorbiaceae, and Fabaceae (Hansford 1961; Hosagoudar 1996; Macedo et al. 2010; Pinho et al. 2012; Pinho et al. 2009; Silva et al. 2012). These host plant families overlap with the host spectrum of sooty mould fungi in the present study. Black mildews have similar traits as the pigmented saprotrophic species, such as the degree of pigmentation and hyphal growth, and can coexist within sooty mould communities. Due to their biotrophic habit, efforts to cultivate black mildews have not been successful (Hansford 1961; Hosagoudar 1996) and they are therefore not included in the species barcoding approach of this study.
At all four sites, sampling efforts were driven by the discovery of dark pigmented epiphytic communities on perennial plant substrates, regardless from the identity of the host plant species. In an earlier study of sooty mould communities on the evergreen Rhododendron ferrugineum, differences in the composition between communities on young leaves and those on previous years leaves were recognised (Flessa and Rambold 2013). A similar effect was observed for the community on annual leaves and perennial structures of Salix spp. in a previous study, and on hibernating evergreens in a greenhouse (Flessa et al. 2012). In the current study design, therefore, only perennial plant parts were compared for all four sites. In the comparative study on sooty mould communities on Salix and evergreen plants in the same region, the influence of different climatic annual cycles was analysed. The sooty mould communities on perennial plant parts of both host groups differed less than the sooty mould communities on annual and perennial structures within one group. As seasonal aspects may have less influence on the composition of sooty mould fungi than on other organisms (Flessa et al. 2012), the chosen study design is suitable to address the hypotheses of the present study. Since sooty moulds were only found on hosts of one genus per site at both European locations, but at the Brazilian sites on host plants of different orders and families, conclusions about host plant dependence can be drawn at least for the two sites in Brazil.
Most current studies do not distinguish between epiphytic and endophytic fungal communities, as the complete microbiome is usually extracted from whole leaves (Lynikiene et al. 2020; Qian et al. 2020; Würth et al. 2019). This is problematic in that endophytic fungi are more host-dependent than epiphytic fungi (Fonseca-García et al. 2016; Lau et al. 2013; Peršoh 2013). Furthermore, it has been shown that the two microbiomes differ considerably and only share a small proportion of the strains (Flessa and Rambold 2013; Yao et al. 2019). In this study, therefore, a methodological approach was chosen that allows to focus on epiphytic communities.
Differences in community composition between continents and sampling sites
The significant difference between the alpha diversity of European and Brazilian sites reflects the distance between the two phytogeographical zones, which differ in climatic conditions and vegetation types. The significant difference in the sooty mould communities can therefore be explained to a considerable extent by climatic differences. In Brazil, the vegetation cycle is usually interrupted by a long dry season with a rise in temperature due to the lack of rain. This has a strong influence on all organisms, comparable to the strong influence of seasonal effects on communities at European sites, but also plants in Europe may be exposed to severe drought in summer and winter (Aulitzky et al. 1982; Cernusca 1976; Rango and Martinec 1994; Sakai and Larcher 1987). However, since the grouping factor ‘continent’ explains the different sooty mould communities less well than the individual sampling sites, the large-scale climatic differences are not the main factor. One possible reason for this overlap could be the dry periods which occur in both sites in Brazil and Europe and may be a stronger driving factor than different average temperatures. Another reason could be that there are generally many ubiquitous fungi in the sooty mould community, and indeed, 5 OTUs occur at all sites.
The different sites (BC, BR, EA, EC) influenced the community composition significantly, with much less overlap between groups. Since all four sites were close to busy roads, the differences cannot be explained by different levels of pollution. Although both European sampling sites are located in the temperate zone, there are significant differences in the type of vegetation surrounding the sampling sites and the mean annual temperature due to the altitude. Climate data from the Botanical Garden weather station at EC showed that the highest amounts of rainfall coincided with the highest temperatures in July and August and with the lowest temperatures in December and January. At EA, the highest amount of precipitation corresponds to the month with the lowest mean temperatures (Tirol Atlas, Geographie Innsbruck). Thus, since the Brazilian fungal communities from Caatinga and rainforest are located at a short distance apart, macroclimatic factors cannot explain these differences. This finding contrasts with the results of Dhami et al. (2013), who found no differences in community composition between two more distant sampling sites with different host plant species, when using a community barcoding approach. In this study, host influence was tested only between plant individuals of two species of the same genus (Nothofagus), but not at higher taxonomic levels. Maybe the differences between two species within a genus are too small to have an influence on the community composition.
Influence of sap-feeding insect association on community composition
There is evidence from many studies that sooty moulds have relationships with leaf-sucking insects because they are able to grow on honeydew (Chomnunti et al. 2014; Perez et al. 2009). However, the presence of honeydew alone does not lead to a defined community composition because significant differences could be observed between communities of EC and BC. Climate and vegetation type are therefore most important, but probably also the fact that honeydew composition greatly differs between Aphididae (EC) and Coccidae (BC). The dominant sugar components in the honeydew of Aphis fabae (Aphididae) are the trisaccharide melezitose and fructose (Fischer et al. 2005), while in Coccus hesperidum (Coccidae), glucose and sucrose are the main sugar components (Golan and Nadja 2011). Different types of sugars (mono-, di-, and trisaccharides) as additional nutrition sources to the substances leached from the leaf tissues could therefore have an indirect influence on the composition of the fungal community. However, the most abundant OTUs were found on plants of both Brazilian sites, although sap-feeding insects were only found on BC host plants. This indicates that the occurrence of those genera is not strictly dependent on the presence of scale insects and honeydew as a nutrition source. Nevertheless, other fungi may be strongly affected by the presence or absence of honeydew in addition to substances leached from the leaf tissues (Tukey 1970). Based on the results of this study, it can be assumed that, although honeydew provides additional carbohydrates for the sooty mould community, it does not lead to a specific sooty mould community as an isolated factor. As the fungi found in sap-feeding insect samples (BC*) largely overlap with those in the sooty mould community on leaves of the same site, these insects could play a role in the dispersal of sooty moulds within a region. It is possible that only a part of the fungi is spread by insects, and others, such as Xylariales, by airborne spores. However, this requires further investigation.
Host-species selectivity
The host plants studied in the tropics and the host plant Rhododendron ferrugineum in the European Alps have perennial leaves. On the European colline site (with mostly deciduous trees), ‘saturated’ epifoliar fungal communities were found on branch tips of Salix spp. and were sampled from there. Thus, at all four sampling sites, premature termination of succession in fungal communities due to leaf fall as a consequence of seasonality can be excluded (Flessa et al. 2012). Host plant order and family had a significant impact on the community composition in both approaches. Differences in the structure of the fungal community among host plant species can be caused by differences in surface structure and nutritional status on leaf surfaces. Leaching substances, which may persist of most compounds found in plants, may serve as other sources than honeydew (Tukey 1970) and vary between different host plant species. This widespread process of leaching leads to large nutrient losses (Schoch 1955; Wallace 1930), so that leaching substances can easily accumulate on the leaf surface of one plant species, but may be swept away from others due to different surface structures.
The assumption that host plant taxonomy has a great influence on phyllosphere fungal community composition is consistent with the study by Kembel and Mueller (2014), who showed that in the tropics the taxonomic identity of the host plant explains more than half of the variation of the composition of fungal communities across trees. The result of the present study is not fully conclusive, however, because of the 15 pairwise tests that included orders from the tropical sites, only 5 were significantly different and among the families only one. This result is not surprising, however, as pathogenic and endophytic fungi are known to be more restricted to certain host plant genera or species, while saprotrophs, which dominate the communities described here, are less host-specific (Kodsueb et al. 2008; Zhou 2001). A large proportion of cultivable fungal species in this study were described as mixotrophic. This two-track nutrition type may be responsible for this relatively low degree of selectivity between host plants. Although host plant order alone does not explain the differences well, five significant results of the pairwise tests of the combined factors ‘host plant order’ and ‘sap-feeding insect association’ from the Brazil sites at least suggest that different leaching in relation to honeydew may have an influence on community composition. However, this would need to be verified in more detail in future studies. Communities on Ericales and Ericaceae from EA differed significantly from orders and families of the other sites. Presence of fungi in the EA samples was mostly restricted to the undersides of the leaves. Leachates or glandular secretions, as present on leaves of Rhododendron ferrugineum (Flessa and Rambold 2013), may provide a substrate leading to a more specific sooty mould community. However, as sooty moulds occurred exclusively on R. ferrugineum in the alpine study site, this cannot be conclusively assessed.
Fungal community structure: taxonomic groups, pigmentation, and nutrition types
The fact that a numerous coexisting species compete for the same resource requires more attention. According to the niche theory, the most common result is competitive exclusion, and it is to be expected that single-species should dominate over oligo-species communities (Tubay et al. 2015). This was observed in Rhododendron-associated fungal communities, where glandular secretions in young leaves were correlated with the dominant occurrence of a sooty mould fungus (Flessa and Rambold 2013). However, this fungus is not able to suppress the growth of other fungi. On perennial leaves, the predominance of that fungal species decreased towards pre- or co-dominance with a ubiquitous species. This result is similar to the situation on evergreen trees sampled in the colline zone (Flessa et al. 2012). The high proportion of oligo-species communities in older Rhododendron ferrugineum leaves, on stems of Salix spp., and on the various leaves of tropical trees thus suggests that, according to the niche theory, more traits added by the community-inhabiting fungi themselves lead to more complex and different niches, which in turn can be filled with additional fungal species.
Capnodiales, common fungi in all four habitats studied, are usually dark pigmented. Capnodiaceae are sooty moulds in the strict sense and their dominance in Brazil is consistent with previous studies on tropical sooty moulds. Together with species of Antennulariellaceae, Capnodiaceae, Chaetothyriaceae, Coccodiniaceae, Euantennariaceae, and Metacapnodiaceae, they dominate the saprotrophic epifoliar fungal community forming sooty mould symptoms (Chomnunti et al. 2014; Dhami et al. 2013; Faull et al. 2002; Olejnik et al. 1999). However, they do not occur in such communities in Europe. There, this niche is occupied by Chaetothyriales sp. (EC) and Capnodiales sp. in coexistence with Sarcinomyces sp. (EA). Of the four OTUs that were ubiquitous at all sites, only Mycosphaerella tassiana (dark pigmented, mixotrophic according to FUNGuild and its cultivability) was also among the more common fungi at all four sites. Less continuous, but present at all sites was Aureobasidium pullulans (mixotrophic, light and dark pigmented).
Fungi which were present in at least two-thirds of all biofilms form the core community of the sooty mould symptom. It is formed by specific, non-ubiquitous and a few ubiquitous fungi. The OTUs, which were present in less than 1/3 of all samples, are considered sporadic companions rather than members of the main matrix in the biofilm of the sooty mould community. A large proportion of these are light pigmented. For this group of fungi, the absence of dark pigmentation on leaf surfaces may be a disadvantage under natural conditions as discussed below. Causes for this lack of pigmentation may also be in vitro effects caused by the type of cultivation media (Lee and Hyde 2002, Lathadevi et al. 2014) so that the proportion of dark pigmented ones could be even higher. Among the common fungi, the proportion of dark pigmented species is lowest in the warmest regions and highest in the colder regions. This is in accordance with the thermal melanism theory (Clusella Trullas et al. 2007) which states that dark pigmented ectothermal organisms have an advantage at low temperatures because they can warm up more easily. Saprotrophic fungi have been subject of a large-scale study in Europe and were found to be darker in cold environments (Krah et al. 2019), which supported the theory of thermal melanism and has now also been confirmed for sooty mould fungi in this study: the proportion of dark pigmented predominant species is lowest in the warmest regions BR (48%) and BC (59%) and higher in the colder regions EC (67%) and EA (100%). Sooty mould communities comprise primary pigmented fungi and accessory unpigmented fungi. The main group consists of functional species that are dark pigmented. Melanin protects against high UV radiation (Zak 2005) and chemical and biological stress, leads to desiccation resistance and structural re-enforcement of hyphal walls (Butler and Day 1998; Eisenman and Casadevall 2012), and leads to the advantage of faster warming in cold regions (Krah et al. 2019). Unpigmented fungi lacking this trait may be considered accessory endomycelial immigrants. A correlation of dark pigmentation with a saprotrophic or mixotrophic nutrition strategy and light pigmentation with a biotrophic strategy was not observed, based on the results of the single-species barcoding approach. It is assumed that both dark pigmented and less or unpigmented species of this fungal community can also live saprotrophically. Some of them may optionally be biotrophic or fungicolous, which is also likely for dark pigmented ones (Butler et al. 2001). In their epifoliar life stage, unpigmented fungi may be dependent on the primary group. The incrustations formed by dark pigmented fungi are thus heterospecific mycelia consisting of primary and secondary fungal components. Biofilms of bacteria of different compositions and structures are well known (Davey and O’Toole 2000; Stahl et al. 2013). They mostly involve semi-aquatic accumulations of prokaryotes. However, there is no reason not to consider heterospecific mycelia as a type of biofilm, according to the definition by Characklis (1990), which refers to ‘cells at a substratum, a surface accumulation, which is not uniform in time or space, a significant amount of inorganic or abiotic substances held together by the biotic matrix and transport and transfer processes play[ing] a much more important role than in isolated occurrence of suspended single cells’ (Characklis 1990). Another indication for different structural niches within the epifoliar fungal community are the different nutritional types. Most species and OTUs (of both approaches) are totally or at least partially saprotrophic. Only 1/5 of all OTUs in this study were identified as biotrophic according to the FUNGuild classification. However, this result has to be put into perspective in that species from the culture-based approach that are classified as biotrophic may only be facultatively biotrophic due to their cultivability. The proportion of possible saprotrophs may be underestimated if the FUNGuild reference alone is used to determine the nutritional type. Of the biotrophic or potentially biotrophic OTUs, the largest proportion was plant-associated with only a small proportion classified as potentially fungicolous.
Conclusions
Based on the results of this study, the core community of sooty moulds can be characterised as follows: Only few species generally dominate and form a main group. Among them, there are usually one or few specific representatives that occur together with a few ubiquitous species with dark pigmentation. Other unpigmented or weakly pigmented fungi are considered accessory and are saprotrophic or biotrophic, the latter interacting directly with their hosts. Their presence depends on the presence of main group representatives that form a protecting structure. In this context, the theory of thermal melanism was supported for the first time for sooty mould communities. Diversity and composition of sooty mould communities depend on site and climatic conditions as well as on the identity of the host plant, but to a lesser extent on the presence of sap-feeding insects. We propose to subsume the heterospecific mycelial complex of sooty mould communities under the term ‘biofilm’. In this study, a first impression could be gained about the potential of comparative mycogeographic studies on habitat and host plant preferences of sooty moulds. Further studies are needed to investigate the influence of sap-feeding insects in more detail.
Availability of data and materials
Sequences from single-species barcoding are available in the NCBI GenBank under accession numbers GU942834–GU942923, KM259875–KM259899, and MN604915-MN604921. Sequences from community barcoding are linked under BioProject accession number PRJNA699866. Reference strains of the single-species community barcoding were deposited in the Jena Microbial Resource Collection (JMRC Jena) as specified under “Material and methods”. Further data cited in the manuscript are available as Supplementary Data. All other relevant data is available upon request.
References
An QL, Ehlers K, Kogel KH et al (2006) Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 172:563–576
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Aulitzky H, Turner H, Mayer H (1982) Bioklimatische Grundlagen einer standortsgemäßen Bewirtschaftung des subalpinen Lärchen-Arvenwaldes. Mitt Eidg Anst Forstl Vers-Wes 58:327–580
Bao L, Cai W, Zhang X et al (2019) Distinct microbial community of phyllosphere associated with five tropical plants on Yongxing Island. South China Sea Microorganisms. https://doi.org/10.3390/microorganisms7110525
Barr ME (1987) Prodomus to class Loculoascomycetes. University of Massachusetts, Amherst
Batista AC, Ciferri R (1963) Capnodiales. Saccardoa 2:1–296
Beng KC, Corlett RT (2019) Identifying the mechanisms that shape fungal community and metacommunity patterns in Yunnan. China Fungal Ecol. https://doi.org/10.1016/j.funeco.2019.08.006
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300
Beule L, Chen K-H, Hsu C-M et al (2019) Soil bacterial and fungal communities of six bahiagrass cultivars. PeerJ. https://doi.org/10.7717/peerj.7014
Blechert O, Zheng H, Zang X et al (2019) Influence of the cultivation medium and pH on the pigmentation of Trichophyton rubrum. PLoS One 14:e0222333. https://doi.org/10.1371/journal.pone.0222333
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Butler M, Day A (1998) Fungal melanins: a review. Can J Microbiol 44(12):1115–1136
Butler MJ, Day AW, Henson JM, Money NP (2001) Pathogenic properties of fungal melanins. Mycologia 93(1):1–8. https://doi.org/10.1080/00275514.2001.12061273
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. https://doi.org/10.1038/ismej.2017.119
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Cernusca A (1976) Structure of forest stand, bioclimatology and energy economy of dwarf shrub communities in Alps. Oecol Plant 11:71–101
Characklis WG (1990) Biofilm processes. In: Characklis WG, Marshall KC (eds) Biofilms. John Wiley & Sons, New York, pp 195–231
Checinska Sielaff A, Urbaniak C, Mohan GBM et al (2019) Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome. https://doi.org/10.1186/s40168-019-0666-x
Chen H (2018) VennDiagram: generate high-resolution Venn and Euler plots. R package version 1(6):20 https://CRAN.R-project.org/package=VennDiagram
Chomnunti P, Hongsanan S, Aguirre-Hudson B et al (2014) The sooty moulds. Fungal Divers 66:1560–2745
Clusella Trullas S, van Wyk JH, Spotila JR (2007) Thermal melanism in ectotherms. J Therm Biol 24:235–245
Davey ME, O’Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867
Dekhuijzen H, Scheer C (1969) The ultrastructure of powdery mildew, Sphaerotheca fuliginea, isolated from cucumber leaves. Eur J Plant Pathol 75:169–177
Del Frari G, Gobbi A, Aggerbeck MR et al (2019) Characterization of the wood mycobiome of Vitis vinifera in a vineyard affected by Esca. Spatial distribution of fungal communities and their putative relation with leaf symptoms. Front Plant Sci 10:910–910. https://doi.org/10.3389/fpls.2019.00910
Dhami MK, Weir BS, Taylor MW, Beggs JR (2013) Diverse honeydew-consuming fungal communities associated with scale insects. PLoS One. https://doi.org/10.1371/journal.pone.0070316
Dissanayake AJ, Purahong W, Wubet T et al (2018) Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers 90:85–107. https://doi.org/10.1007/s13225-018-0399-3
Egidi E, Delgado-Baquerizo M, Plett JM et al (2019) A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun. https://doi.org/10.1038/s41467-019-10373-z
Eisenman HC, Casadevall A (2012) Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol 55:412–418
Epstein HE, Torda G, van Oppen MJH (2019) Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 38:373–386. https://doi.org/10.1007/s00338-019-01783-y
Ezeokoli OT, Mashigo SK, Maboeta MS et al (2020) Arbuscular mycorrhizal fungal community differentiation along a post-coal mining reclamation chronosequence in South Africa: a potential indicator of ecosystem recovery. Appl Soil Ecol. https://doi.org/10.1016/j.apsoil.2019.103429
Faull JL, Olejnik I, Ingrouille M, Reynolds D (2002) A reassessment of the taxonomy of some tropical sooty molds. Trop Mycol 2:33–40
Fischer M, Völkl W, Hoffmann KH (2005) Honeydew production and honeydew sugar composition of polyphagous black bean aphid, Aphis fabae (Hemiptera: Aphididae) on various host plants and implications for ant-attendance. Eur J Entomol 105:155–160
Flessa F, Rambold G (2013) Diversity of the Capnocheirides rhododendri-dominated fungal community in the phyllosphere of Rhododendron ferrugineum L. Nova Hedwigia 97:19–53
Flessa F, Kehl A, Kohl M (2010) RFLPtools: tools to analyse RFLP data. R package version 1:6 https://cran.r-project.org/web/packages/RFLPtools/index.html
Flessa F, Peršoh D, Rambold G (2012) Annuality of Central European deciduous tree leaves delimits community development of epifoliar pigmented fungi. Fungal Ecol 5:554–561
Folmer O, Black M, Hoeh W et al (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fonseca-García C, Coleman-Derr D, Garrido E et al (2016) The cacti microbiome: interplay between habitat-filtering and host-specificity. Front Microbiol 7:150–150. https://doi.org/10.3389/fmicb.2016.00150
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Golan K, Nadja A (2011) Differences in the sugar composition of the honeydew of polyphagous brown soft scale Coccus hesperidum (Hemiptera: Sternorrhyncha: Coccoidea) feeding on various host-plants. Eur J Entomol 108:705–709
Hamid M, Jalaluddin M (2006) Recurring incidence of sooty mould of mango in Karachi and its control. Int J Biol Biotechnol 3:561–565
Hansford CG (1961) The Meliolineae. Sydowia Ann Mycol Ser. II, Beihefte 2:1–806
Hollingsworth ML, Clark AA, Forrest LL et al (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457
Hosagoudar V (1996) Meliolales of India. Bot Surv India Calcutta 1:363
Hughes SJ (1976) Sooty molds. Mycologia 68:693–820
Janakiev T, Dimkić I, Unković N et al (2019) Phyllosphere fungal communities of plum and antifungal activity of indigenous phenazine-producing Pseudomonas synxantha against Monilinia laxa. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02287
Janowsky J, Kimbrough E, Kandalepas D et al (2019) Bacterial and fungal endophyte communities differ in trees of natural versus wastewater-treatment wetlands. Wetl Ecol Manag 27:711–723. https://doi.org/10.1007/s11273-019-09688-x
Kandlikar G (2019) ranacapa: Utility functions and “shiny” app for simple environmental DNA visualizations and analyses. Version R package version 0.1.0. https://github.com/gauravsk/ranacapa
Kembel SW, Mueller RC (2014) Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92:303–311
Kirk PM, Cannon PF, Minter DW, Stalpers JA (eds) (2010) Dictionary of the fungi, 10th Edition, 10th edn. CABI, Wallingford
Kodsueb R, McKenzie EHC, Lumyong S, Hyde KD (2008) Diversity of saprobic fungi on Magnoliaceae. Fungal Divers 30:37–53
Kolde R (2019) pheatmap: pretty heatmaps. Version R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap
Kowalski T, Nawrot-Chorabik K, Grad B, Leszczyński K (2016) Influence of culture conditions on medium discoloration and mycelial growth of Dothistroma septosporum. For Pathol 46:507–514. https://doi.org/10.1111/efp.12243
Krah FS, Büntgen U, Schaefer H et al (2019) European mushroom assemblages are darker in cold climates. Nat Commun 10:2890. https://doi.org/10.1038/s41467-019-10767-z
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lathadevi KC, Chandi CR, Bapuji M, Gulnihal O (2014) Culture conditions for growth and pigment production of mangrove Penicillium species. J Multidiscip Sci Res 2:1–5
Lathi L, Shetty S (2017) Microbiome R package: tools for microbiome analysis in R. Version 1.9.19. http://microbiome.github.com/microbiome
Lau MK, Arnold AE, Johnson NC (2013) Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol 6:365–378. https://doi.org/10.1016/j.funeco.2013.06.003
Lee OHK, Hyde KD (2002) Phylloplane fungi in Hong Kong mangroves: evaluation of study methods. Mycologia 94:596–606
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:256–259. https://doi.org/10.1093/nar/gkz239
Li P-D, Jeewon R, Aruna B et al (2019) Metabarcoding reveals differences in fungal communities between unflooded versus tidal flat soil in coastal saline ecosystem. Sci Total Environ 690:911–922. https://doi.org/10.1016/j.scitotenv.2019.06.473
Lynikiene J, Marciulyniene D, Marciulynas A et al (2020) Managed and unmanaged Pinus sylvestris forest stands harbour similar diversity and composition of the phyllosphere and soil fungi. Microorganisms. https://doi.org/10.3390/microorganisms8020259
Macedo DM, Pinho DB, Barreto RW et al (2010) Black mildew fungi (Meliolaceae) associated with Schinus terebinthifolius (Brazilian pepper tree) in Brazil. Mycotaxon 114:429–437
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. https://doi.org/10.14806/ej.17.1.200
Martinez P (2019) pairwiseAdonis: pairwise multilevel comparison using Adonis. Version R package version 0.3. https://github.com/pmartinezarbizu/pairwiseAdonis
Mbareche H, Veillette M, Bilodeau G, Duchaine C (2020) Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ. https://doi.org/10.7717/peerj.8523
McMurdie PJ, Holmes S (2013) phyloseq: A=an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. https://doi.org/10.1371/journal.pone.0061217
Morales-Rodriguez C, Sferrazza I, Aleandri M et al (2019) Fungal community associated with adults of the chestnut gall wasp Dryocosmus kuriphilus after emergence from galls: taxonomy and functional ecology. Fungal Biol 123:905–912. https://doi.org/10.1016/j.funbio.2019.09.009
Nelson S (2008) Sooty mold. Plant Dis 52:1–6
Nerva L, Pagliarani C, Pugliese M et al (2019) Grapevine phyllosphere community analysis in response to elicitor application against powdery mildew. Microorganisms 7:662. https://doi.org/10.3390/microorganisms7120662
Neuwirth E (2014) RColorBrewer: ColorBrewer Palettes. Version R package version 1:1–2 https://CRAN.R-project.org/package=RColorBrewer
Nguyen N, Song Z, Bates S et al (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Nilsson RH, Larsson KH, Taylor A et al (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Oksanen J, Blanchet G, Friendly M et al (2019) vegan: community ecology package. Version 2:5–6 https://CRAN.R-project.org/package=vegan
Olejnik IM, Ingrouille M, Faull JL (1999) Numerical taxonomy of the sooty moulds Leptoxyphium, Caldariomyces and Aithaloderma based on micromorphology and physiology. Mycol Res 103:333–346
Palacio-Barrera AM, Areiza D, Zapata P et al (2019) Induction of pigment production through media composition, abiotic and biotic factors in two filamentous fungi. Biotechnol Rep 21:e00308. https://doi.org/10.1016/j.btre.2019.e00308
Pan Y, Wu Y, Li X et al (2019) Continuing impacts of selective inhibition on bacterial and fungal communities in an agricultural soil. Microb Ecol 78:927–935. https://doi.org/10.1007/s00248-019-01364-0
Parbery IH, Brown JF (1986) Sooty moulds and black mildews in extratropical rainforests. In: Fokkema NJ, Van den Heuvel J (eds) Microbiology of the phyllosphere. Cambridge University Press, London, pp 101–120
Perez JL, French JV, Summy KR et al (2009) Fungal phyllosphere communities are altered by indirect interactions among trophic levels. Microb Ecol 57:766–774
Peršoh D (2013) Factors shaping community structure of endophytic fungi–evidence from the Pinus-Viscum-system. Fungal Divers 60:55–69. https://doi.org/10.1007/s13225-013-0225-x
Peršoh D, Melcher M, Flessa F, Rambold G (2010) First fungal community analyses of endophytic ascomycetes associated with Viscum album ssp. austriacum and its host Pinus sylvestris. Fungal Biol 114:585–596
Pietrowski A, Flessa F, Rambold G (2010) Towards an efficient phenotypic classification of fungal cultures from environmental samples using digital imagery. Mycol Prog 11:383–393
Pinho DB, Pereira OL, Nicoli A et al (2009) Black mildew disease on the neotropical medicinal shrub Pothomorphe umbellata in Brazil, caused by Irenopsis tortuosa var. potomorphes. Australas Plant Dis Notes 4:766–774
Pinho DB, Pereira OL, Firmino AL et al (2012) New Meliolaceae from the Brazilian Atlantic forest 1. Species on hosts in the families Asteraceae, Burseraceae, Euphorbiaceae, Fabaceae and Sapindaceae. Mycologia 104:121–137
Qian X, Li S, Wu B et al (2020) Mainland and island populations of Mussaenda kwangtungensis differ in their phyllosphere fungal community composition and network structure. Sci Rep 10:952. https://doi.org/10.1038/s41598-020-57622-6
R Core Team (2020) R: A language and environment for statistical computing. Version 3.6.2. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Rango A, Martinec J (1994) Areal extent of seasonal snow cover in a changed climate. Nord Hydrol 25:33–52
Ricks KD, Koide RT (2019) The role of inoculum dispersal and plant species identity in the assembly of leaf endophytic fungal communities. PLoS One. https://doi.org/10.1371/journal.pone.0219832
Rodríguez D, Piepenbring M (2007) Two new species of Appendiculella (Meliolaceae) from Panama. Mycologia 99:544–552
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ. https://doi.org/10.7717/peerj.2584
Roskov Y, Ower T, Orrell T, et al. (eds) (2019) Species 2000 & ITIS Catalogue of Life, 2019 Annual Checklist. Naturalis, Leiden, the Netherlands
Saddhe AA, Jamdade RA, Kumar K (2017) Evaluation of multilocus marker efficacy for delineating mangrove species of West Coast India. PLoS One. https://doi.org/10.1371/journal.pone.0183245
Sakai A, Larcher W (1987) Frost survival of plants. Responses and adaption of freezing stress. In: Billings WD, Golley F, Lange OL et al (eds) Ecological studies. Springer-Verlag, Berling, p 321
Saravesi K, Markkola A, Taulavuori E et al (2019) Impacts of experimental warming and northern light climate on growth and root fungal communities of Scots pine populations. Ecol Mycorrhizas Anthr 40:43–49. https://doi.org/10.1016/j.funeco.2018.12.010
Schiro G, Colangeli P, Muller MEH (2019) A metabarcoding analysis of the mycobiome of wheat ears across a topographically heterogeneous field. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02095
Schoch K (1955) Quantitative determinations of the cuticular secretion of K and Ca. Ber Schweiz Bot Ges 65:205–250
Schoulties CL (1980) Sooty molds. Plant Pathol Circ 208:1–2
Silva M, Barreto RW, Pereira OL (2012) Fungal pathogens of ‘cat’s claws’ from Brazil for biocontrol of Macfadyena unguis-cati. Mycotaxon 119:181–195
Simon C, Frati F, Beckenbach A et al (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Stahl DA, Flowers JJ, Hullar M, Davidson S (2013) Structure and function of microbial communities. In: Rosenberg E, Delong EF, Lory S et al (eds) The prokaryotes, 4th edn. Springer References, pp 3–30
Triebel D, Peršoh D, Wollweber H, Stadler M (2005) Phylogenetic relationships among Daldinia, Entonaema, and Hypoxylon as inferred from ITS nrDNA analyses of Xylariales. Nova Hedwigia 80:25–43
Tubay JM, Suzuki K, Uehara T et al (2015) Microhabitat locality allows multi-species coexistence in terrestrial plant communities. Sci Rep 5:15376. https://doi.org/10.1038/srep15376
Tukey HB (1970) Leaching of substances from plants. Annu Rev Plant Physiol 21:305–324
Wallace T (1930) Experiments on the effects of leaching with cold water on the foliage of fruit trees.: I. The course of leaching of dry matter, ash and potash from leaves of apple, pear, plum, black currant and gooseberry. Jour Pomol Hort Sci 8:44–60. https://doi.org/10.1080/03683621.1930.11513349
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc., San Diego, New York, Berkeley, Boston, London, Sydney, Tokyo, Toronto, p 482
Whittaker RH (1960) Vegetation of the Siskiyou Mountains, Oregon and California. Ecol Monogr 30:279–338
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag New York
Wilkinson TDJ, Miranda J-P, Ferrari J et al (2019) Aphids influence soil fungal communities in conventional agricultural systems. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00895
Würth DG, Dahl MB, Trouillier M et al (2019) The needle mycobiome of Picea glauca – a dynamic system reflecting surrounding environment and tree phenological traits. Fungal Ecol 41:177–186. https://doi.org/10.1016/j.funeco.2019.05.006
Yao H, Sun X, He C et al (2019) Phyllosphere epiphytic and endophytic fungal community and network structures differ in a tropical mangrove ecosystem. Microbiome. https://doi.org/10.1186/s40168-019-0671-0
Zak J (2005) Fungal communities of deserts ecosystems. Link to climate change. In: Dighton J, White JF, Oudemans P (eds) The fungal community. Its organization and role in the ecosystem, 3rd edn. CRC Press, pp 659–681
Zhang Z, Kong X, Jin D et al (2019) Euonymus japonicus phyllosphere microbiome is significantly changed by powdery mildew. Arch Microbiol 201:1099–1109. https://doi.org/10.1007/s00203-019-01683-3
Zhou D (2001) Host-specificity, host-exclusivity, and host-recurrence in saprobic fungi. Mycol Res 105:1449–1457
Acknowledgements
We acknowledge Dr. Andreas Brachmann, Biocenter of LMU (Munich) for Illumina sequencing and Christina Leistner (Bayreuth) for preparative work in the laboratory.
Funding
Open Access funding enabled and organized by Projekt DEAL. The visit of the collection sites in the Mata Atlântica relic rainforest in October 2013 was funded by the DFG (RA 731/15-1).
Author information
Authors and Affiliations
Contributions
Fabienne Flessa co-designed the project, collected the environmental samples from Europe, carried out part of the laboratory work as well as all the statistical analyses in this study, and wrote the original draft of the manuscript taking into account the input of the co-authors. Janno Harjes wrote sections on methodology and bioinformatics, carried out part of the laboratory work, and established and run the bioinformatic pipeline for community barcoding. Marcela Cáceres collected the environmental samples from Brazil. Gerhard Rambold designed and supervised the project, revised the manuscript, and collected and provided the environmental samples from Brazil.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human or animal subjects
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Claus Bässler
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised. Supplementary file has been added.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Flessa, F., Harjes, J., Cáceres, M.E.S. et al. Comparative analyses of sooty mould communities from Brazil and Central Europe. Mycol Progress 20, 869–887 (2021). https://doi.org/10.1007/s11557-021-01700-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01700-0