Abstract
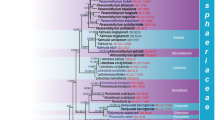
The phylogenetic relationships of five morphologically similar strains labeled Exochalara longissima were studied with sequences of Internal Transcribed Spacer (ITS1-nc5.8S-ITS2) and the small and large subunits of nuclear ribosomal DNA (nc18S and nc28S rDNA) in three different molecular data sets. The phylogenetic analyses, maximum likelihood, maximum parsimony heuristic search and Bayesian approach, revealed that these strains belong to three different genera. Based on the two-gene phylogeny of nc18S-nc28S rDNA, the true relationship of the anamorphic genus Exochalara, typified with E. longissima (strains CBS 393.82 and CBS 980.73), lies within the Helotiales of the Leotiomycetes. Exochalara is characterized by upright, pigmented conidiophores with terminally integrated cylindrical monophialides and hyaline fusiform to drop-shaped conidia, cohering end-to-end in basipetal chains. The holomorph genera Hyphodiscus (anamorph Catenulifera) and Chlorociboria (anamorph Dothiorina) are shown as closest relatives of Exochalara. The strains CBS 622.82 and CBS 114633, which differ from Exochalara in shorter, decumbent conidiophores, ampulliform monophialides, ellipsoidal to obovoidal, 1-celled hyaline conidia straw-yellow in mass and sympodial ramification/branching of conidiophores in vitro also grouped in the Leotiomycetes but on a separate position from Exochalara. The new genus Brachyalara is introduced for them. Subsequently four strains of the morphologically similar Chalara microchona were analysed. Three strains, including the ex-type strain, which formed a strongly supported clade distinct from more typical species of Chalara, Brachyalara and Exochalara, are introduced as a new monotypic genus Infundichalara. Exochalara and the two newly recognized genera are compared with Chalara, Herreromyces and Phialophora. The strain MUCL 40959, according to the ITS (ITS1-ITS2) and three-gene phylogeny (nc18S-nc5.8S-nc28S rDNA) is found to belong to Monilochaetes, which is the anamorph of the holomorph genus Australiasca in the Glomerellales, Sordariomycetes. The fungus is described as a new anamorphic species and compared with the other four species in the genus.









Similar content being viewed by others
References
Barr ME, Blackwell M (1980) A new genus of the Lophiaceae. Mycologia 72:1224–1227
Berthet P (1974) Formes conidiennes de divers Discomycètes. Bull Trimest Soc Mycol Fr 80:125–149
Blackwell M, Gilbertson RL (1985) Quasiconcha reticulata and its anamorph from conifer roots. Mycologia 77:50–54
Bogale M, Orr MJ, O’Hara MJ, Untereiner WA (2010) Systematics of Catenulifera (anamorphic Hyaloscyphaceae) with an assessment of the phylogenetic position of Phialophora hyalina. Fung Biol 114:396–409
Cai L, Wu WP, Hyde KD (2009) Phylogenetic relationships of Chalara and allied species inferred from ribosomal DNA sequences. Mycol Prog 8:133–143
Castañeda RF, Kendrick BW (1991) Ninety-nine conidial fungi from Cuba and three from Canada. Univ Waterloo Biol Ser 35:1–132
Constantinescu O, Holm K, Holm J (1995) Teleomorph-anamorph connections in Ascomycetes: the anamorphs of three species of Chaetosphaeria. Mycol Res 99:585–592
Crous PW, Groenewald (Ewald) JZ, Gams W (2003) Eyespot of cereals revisited: ITS phylogeny reveals new species relationships. Eur J Pl Path 109:841–850
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Mol Phyl Evol 14:733–740
Damm U, Fourie PH, Crous PW (2010) Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 24:60–80
de Hoog GS, Weenink XO, Gerrits van den Ende AH (1999) Taxonomy of the Phialophora verrucosa complex with the description of two new species. Stud Mycol 43:107–122
Dixon JR (1975) Chlorosplenium and its segregates. II. The genera Chlorociboria and Chlorencoelia. Mycotaxon 1:193–237
Ellis MB (1976) More dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey, England
Gams W (1973) Phialides with solitary conidia? Remarks on conidium ontogeny in some Hyphomycetes. Persoonia 7:161–169
Gams W (2000) Phialophora and some morphologically little-differentiated anamorphs of divergent ascomycetes. Stud Mycol 45:187–199
Gams W, Holubová-Jechová V (1976) Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Stud Mycol 13:1–99
Gams W, Hoekstra ES, Aptroot A (eds) (1998) CBS course of mycology, 4th edn. Centraalbureau voor Schimmelcultures, Baarn
Gargas A, Taylor JW (1992) Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 84:589–592
Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V, Fraker E, Schoch CL, Tibell L, Untereiner WA, Aptroot A (2006) Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 98:1053–1064
Gutell RR (1993) Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nuc Acids Res 21:3051–3054
Gutell RR, Gray MW, Schnare MN (1993) A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nuc Acids Res 21:3055–3074
Hall TA (1999) BioEdit 5.0.9: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symp Ser 41:95–98
Harrington TC, McNew DL (2003) Phylogenetic analysis places the phialophora-like anamorph genus Cadophora in the Helotiales. Mycotaxon 87:141–151
Hosoya T (2002) Hyaloscyphaceae in Japan (6): the genus Hyphodiscus in Japan and its anamorph Catenulifera gen. nov. Mycoscience 43:47–57
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Jacobs A, Coetzee MPA, Wingfield BD, Jacobs K, Wingfield MJ (2003) Phylogenetic relationships among Phialocephala species and other ascomycetes. Mycologia 95:637–645
Kauff F, Lutzoni F (2002) Phylogeny of the Gyalectales and Ostropales (Ascomycota, Fungi): among and within order relationships based on nuclear ribosomal RNA small and large subunits. Mol Phyl Evol 25:138–156
Kirk PM, Spooner BM (1984) An account of the fungi of Arran, Gigha and Kintyre. Kew Bull 38:503–597
Kornerup A, Wanscher JH (1978) Methuen handbook of colour, 3rd edn. Eyre Methuen, London
Landvik S (1996) Neolecta, a fruit-body-producing genus of the basal ascomycetes, as shown by SSU and LSU DNA sequences. Mycol Res 100:199–202
Larget B, Simon DL (1999) Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol 16:750–759
Miller AN, Huhndorf SM (2004) A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycol Res 108:26–34
Nag Raj TR, Kendrick WB (1975) A monograph of Chalara and allied genera. Wilfr. Laurier Univ Press, Waterloo
Nag Raj TR, Kendrick WB (1993) The anamorph as generic determinant in the holomorph: The Chalara connection in the Ascomycetes, with special reference to the Ophiostomatoid fungi. In: Wingfield MJ, Seifert KA, Webber JF (eds) Ceratocystis and Ophiostoma. Taxonomy, Ecology and Pathogenicity, pp 61–70
Nixon KC, Carpenter JM (1993) On outgroups. Cladistics 9:413–426
Nylander J (2008) MrModeltest2 v. 2.3. C program for selecting DNA substitution models using PAUP*
Paulin AE, Harrington TC (2000) Phylogenetic placement of anamorphic species of Chalara among Ceratocystis species and other ascomycetes. Stud Mycol 45:209–222
Paulin-Mahady AE, Harrington TC, McNew DL (2002) Phylogenetic and taxonomic evaluation of Chalara, Chalaropsis, and Thielaviopsis anamorphs associated with Ceratocystis. Mycologia 94:62–72
Réblová M, Winka K (2000) Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycologia 92:939–954
Réblová M, Gams W, Seifert KA (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud Mycol (In Press)
Rehner S, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634
Rong IH, Gams W (2000) The hyphomycete genera Exochalara and Monilochaetes. Mycotaxon 76:451–462
Seifert KA (1989) Coryne trichophora comb. nov., and the implications of its conidial anatomy. Stud Mycol 31:157–164
Sivanesan A, Alcorn JL (2002) Australiasca queenslandica gen. et sp. nov. (Chaetosphaeriaceae: Ascomycota) and its anamorph Dischloridium camelliae sp. nov. from Australia. Aust J Bot 15:741–747
Spatafora JW, Mitchell TG, Vilgalys R (1995) Analysis of genes coding for small-subunit rRNA sequences in studying phylogenetics of dematiaceous fungal pathogens. J Clin Microbiol 33:1322–1326
Spatafora JW, Sung GH, Johnson D, Hesse C, O'Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, Reeb V, Gueidan C, Fraker E, Lumbsch T, Lucking R, Schmitt I, Hosaka K, Aptroot A, Roux C, Miller AN, Geiser DM, Hafellner J, Hestmark G, Arnold AE, Budel B, Rauhut A, Hewitt D, Untereiner WA, Cole MS, Scheidegger C, Schultz M, Sipman H, Schoch CL (2007) A five-gene phylogeny of Pezizomycotina. Mycologia 98 (2006) :1018–1028
Stamatakis A, Ludwig T, Meier H (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates
Untereiner WA, Naveau FA (1999) Molecular systematics of the Herpotrichiellaceae with an assessment of the phylogenetic positions of Exophiala dermatitidis and Phialophora americana. Mycologia 91:67–83
Untereiner WA, Naveau FA, Bachewich J, Angus A (2006) Evolutionary relationships of Hyphodiscus hymeniophilus (anamorph Catenulifera rhodogena) inferred from β-tubulin and nuclear ribosomal DNA sequences. Can J Bot 84:243–253
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Vilgalys R, Sun BL (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci USA 91:4599–4603
Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS (2007) Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98 (2006) :1065–1075
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Wuyts J, van de Peer Y, Winkelmans T, de Wachter R (2002) The European database on small subunit ribosomal RNA. Nuc Acids Res 30:183–185
Acknowledgement
This study was supported by the Project of the National Foundation of the Czech Republic (GA206/09/0547) and Institutional Research Concepts No. AV0Z60050516 (Institute of Botany of the ASCR) and No. AV0Z50200510 (Institute of Microbiology of the ASCR). We are very grateful to Dr. R.F. Castañeda-Ruíz, who sent us permanent slides of Monilochaetes dimorphospora, Dr. C. Decock, Prof. P. Crous and the curators of the CBS culture collection for supplying reference strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Réblová, M., Gams, W. & Štěpánek, V. The new hyphomycete genera Brachyalara and Infundichalara, the similar Exochalara and species of ‘Phialophora sect. Catenulatae’ (Leotiomycetes). Fungal Diversity 46, 67–86 (2011). https://doi.org/10.1007/s13225-010-0077-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-010-0077-6