Abstract
Scedosporium and Pseudallescheria fungi are increasingly encountered fungal pathogens. Species identification is important as there are differences in epidemiology, virulence, and antifungal susceptibility that vary according to species or species groups. Histological and culture techniques are limited by low sensitivity and specificity but improved culture approaches employing selective media have improved the recovery of these fungi from clinical samples. Molecular- and proteomics-based methods are increasingly explored and used for species identification. These include multiplex polymerase chain reaction (PCR) and other PCR-based methods, oligonucleotide arrays, DNA sequencing, rolling circle amplification, and matrix-assisted laser desorption time-of-flight mass spectrometry. These techniques have been harnessed for the detection and identification of Scedosporium and Pseudallescheria species directly from clinical samples and from cultures. This review summarizes the methods currently used to enable an informed choice of detection and/or identification techniques in the clinical mycology laboratory.
Similar content being viewed by others
Introduction
Fungi of the genera Scedosporium and Pseudallescheria are important pathogens, especially in immunocompromised stem cell and solid organ transplant recipients [1–3]. They are of concern due to the high mortality of resultant infection and their resistance to many antifungal agents [4–6]. In the immunocompromised, fever and disseminated infection, including central nervous system (CNS), lung and cutaneous disease, are common (reviewed by Cortez et al. [7•] and Husain et al. [8]). Since these fungi are ubiquitous in soil and polluted water, infection may also be acquired through inhalation (including near-drowning accidents) or inoculation of the organism from trauma [7•, 9] with wide-ranging manifestations, including brain abscess, mycetoma, and osteomyelitis. Scedosporium/Pseudallescheria can colonize damaged airways [7•] and this colonization may persist and later result in invasive disease [10, 11].
Scedosporium infections are often clinically indistinguishable from other invasive mould infections and diagnosis relies on recovery or detection of the etiological agent from clinical specimens; however, histopathological findings are non-specific and culture is insensitive. Serological methods, such as immunodiffusion, are either not commercially available or cross-reactions occur with other fungi (summarized by Cortez et al. [7•] and Guarro et al. [12]). In addition, species identification of these fungi is challenged by their evolving taxonomy. While previously comprising two species of clinical interest—Scedosporium prolificans and Scedosporium apiospermum (previous anamorph of Pseudallescheria boydii)—the S. apiospermum/P. boydii complex now encompasses at least eight species and P. boydii and S. apiospermum have been separated as two distinct species [13•, 14, 15]; the most frequent species among clinical isolates are S. apiospermum, S. aurantiacum and P. boydii, [13•, 16]. However, the prevalence of various species remains uncertain due to the inability of many routine mycological methods to fully recognize them. Indeed, it is anticipated that the nomenclature of these fungi will continue to evolve. Identification is important because of species-specific differences in epidemiology, antifungal susceptibility and, possibly, virulence [4, 7•, 16•, 17, 18].
To address current limitations in laboratory diagnosis, numerous molecular- and proteomic-based methods have been developed and are being introduced into the diagnostic laboratory. Moreover, standard culture-based mycological techniques have undergone refinement to increase the yield of Scedosporium/Pseudallescheria from clinical specimens. In this review, we focus on the challenges facing both traditional mycological and more novel diagnostic methods of scedosporiosis caused by the major medically important species. Recovery of these fungi should always be interpreted in combination with clinical findings.
Microscopy and Culture
Despite suboptimal sensitivity and slow turnaround times (2–7 days), detection and recovery by culture of Pseudallescheria and Scedosporium species from clinical specimens remains a cornerstone of the diagnosis of scedosporiosis. These fungi can be recovered from any specimen, including blood culture, skin and soft tissue, bone and tissue biopsy specimens; however, in practice they often present in sputum and other respiratory tract samples. All isolations from sterile sites are considered clinically significant.
Histopathology Stains
Visualization of fungal hyphae in tissue (and other) specimens by traditional Grocott methenamine silver (GMS) or periodic acid-Schiff (PAS) staining is essential to the diagnosis of invasive scedosporiosis since these moulds can colonize airways. Indeed, demonstration of fungal hyphae in tissue is a requirement of the European Organization for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) classification for a ‘proven’ fungal infection (versus colonization) [19].
However, the histopathological appearance of members of the Pseudallescheria/Scedosporium species complex in tissue sections and cytological smears is very similar to Aspergillus spp., Fusarium spp. and other hyphomycete fungi, leading to frequent misidentification [7•, 20]. Some experts testify that in contrast to the regular dichotomous branching pattern typical of Aspergillus hyphae, Scedosporium hyphae are slightly more irregular, with multiple branching at acute angles, providing a distinguishing characteristic [20, 21]; however, in practice, this was found to have low discriminatory value. Others reported unique histopathological characteristics observed, which include parallel hyphae bridging at right angles, in H patterns, and intravascular conidiation and conidia present in tissue are terminal or intercalary globose chlamydospores [7•, 21, 22]. An experienced histopathologist is required to observe these distinctive features.
Microscopic examination of brain abscess specimens due to Scedosporium species have revealed mostly granulomatous inflammation with multinucleate cellular tissue reaction and hyphae penetrating the cerebral blood vessels [23, 24]. In situ hybridization (ISH) techniques using specific fluorescent antibody to bind fungal elements have also been developed to differentiate Aspergillus and Scedosporium in tissue sections [20, 25]; however, the use of such immunohistochemistry approaches has not yet been validated for diagnostic purposes.
Culture and Identification
Scedosporium and Pseudallescheria species will grow on standard mycological media such as Sabouraud’s dextrose agar (SDA); however, their recovery from clinical specimens may be overlooked or masked, especially if there is concurrent growth of fast-growing fungi such as Aspergillus, Fusarium and Candida species [26, 27], leading to underestimation of their presence and frequency. To address this limitation, more effective isolation and recovery can be achieved with the use of semi-selective isolation media. For instance, agents such as dichloran (2,6-dichloro-4-nitroaniline), benomyl, penicillin, streptomycin, chloramphenicol, and cycloheximide are effective at reducing growth of non-Scedosporium moulds, including Aspergillus or Candida spp. [28, 29]. Currently, one of the more efficient media for selective isolation of Scedosporium species is Scedosporium Selective agar (SceSel+), which contains the antifungal agents benomyl and dichloran that inhibit the fast growth of Candida and Aspergillus species [27, 29, 30•]. Other effective semi-selective media include Dichloran Rose Bengal Chloramphenicol (DRBC; Thermo-Scientific/Oxoid, UK) containing benomyl [13•, 31], which has the added advantage of being commercially available. Another medium Mycosel™ agar (BBL, Becton Dickinson (BD), USA) containing chloramphenicol and cycloheximide was found to be inferior to SceSel+ agar for the recovery of Scedosporium from the sputa of cystic fibrosis patients [30•]. SceSel + media is not available commercially and must, ideally, be freshly prepared. It should be noted that, unlike other Scedosporium/Pseudallescheria species, S. prolificans does not grow on media containing cycloheximide (actidione) [see Table 1].
The combination of semi-selective media, i.e. SceSel+ agar or DRBC medium, and standard mycological media, e.g. SDA or potato dextrose agar (PDA), is currently recommended for the recovery of Scedosporium and Pseudallescheria species [30•]. Generally, these species begin to grow at 2 days and are then subcultured onto PDA plates for macroscopic identification and for slide cultures with staining with lacto-phenol cotton blue (LPCB) for microscopic identification. Primary plates should be kept for 10–14 days [30•] and inspected every second day. The sexual stages (Pseudallescheria) are induced using PDA or cornmeal (CM) agar, both as a slide culture and after extended incubation.
Macroscopic Features
Members of the Pseudallescheria/Scedosporium complex all grow moderately fast on PDA at 25 ºC, and mature in approximately 1 week [7•]. Table 1 summarizes the more pertinent macroscopic and colony color characteristics of the medically important species.
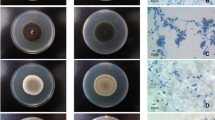
Colonies of S. aurantiacum are dense, cottony to lanose (woolly), frequently with a concentric growth pattern of aerial mycelium, varying from yellow to grey or brown, and usually with a whitish irregularly lobate and fimbriate margin. The reverse is brown-orange at the center and brown to colorless towards the periphery. A distinctive characteristic of S. aurantiacum is evidence of a yellow diffusible pigment on PDA after a few days incubation (Table 1) [13•] (see Fig. 1a). In contrast, S. prolificans colonies begin as a downy colony, which becomes increasingly cottony, with an olive-grey to black coloration (Table 1). The reverse appears dark brown or grey to almost black (Fig. 1c).
The panels illustrate the culture morphology on PDA at 25 ºC comprising plate obverse and reverse, and microscope morphology after staining with lacto-phenol cotton blue (magnification 400×). (a) Scedosporium aurantiacum, (b) Scedosporium apiospermum, (c) Scedosporium prolificans, (d) Scedosporium dehoogii, and (e) Scedosporium boydii/Pseudallescheria boydii. PDA potato dextrose agar
S. apiospermum and P. boydii colonies are initially white to grey, later becoming dark grey or smoky brown, and are woolly to cottony in texture. From the reverse, the color is pale with brownish black zones (Figs. 1c and e). These species are unable to be distinguished from each other by colony morphology [14].
On PDA, Scedosporium dehoogii colonies are cottony white to pale grey with a colorless reverse (Fig. 1d). Colonies grow slowly at 37 °C, reaching only 5–10 mm in diameter after 14 days [14]. The macroscopic characteristics of P. minutispora are summarized in Table 1 [13•].
Microscopic Features
Speciation of Scedosporium/Pseudallescheria species on the basis of microscopic features is reliant on considerable expertise and can be problematic. All isolates can be identified by the presence of a characteristic Scedosporium anamorph. The development of a Graphium synanamorph may or may not be present, not being exclusive to any species (Table 2). It is characterized by the production of synnemata terminating in a slimy head of conidia, varying in size from 80–750 μm long. The teleomorph state (where known) and characteristics of ascospores and conidia for the major Scedosporium species are shown in Table 2.
Microscopically, S. aurantiacum (teleomorph unknown) is characterized by solitary conidiophores on aerial mycelium, which are often reduced to a conidiogenous cell growing percurrent, laterally or terminal from an undifferentiated mycelium. Conidia are typically smooth and thick-walled, obovoid or subcylindrical [7•, 13•]. S. prolificans hyphae are hyaline and septate from which conidiogenous cells are produced along the length; flask-shaped conidiophores are distinctive, which are basally swollen (inflated) from which single, or clusters of, conidia emerge (Fig. 1; Table 2). The ovoid to pyriform conidia are hyaline to pale brown [7•].
P. boydii is homothallic, producing brown cleistothecia, 100–300 μm in diameter. The ascocarp wall is a single layer of thin, flat, polygonal, jigsaw-shaped brown cells, which at maturation burst to release asci filled with ascospores [7•]. These asci are subglobose to globose and contain eight unicellular ascospores, which are pale yellow or brown, smooth, and ovo-ellipsoidal. These sexual ascospores may be distinguished from the asexually-generated conidia by the presence of an internal oil droplet and absence of a truncated base [7•]. S. apiospermum and P. boydii, although two seprate species, cannot be separated by microscopic features [12, 13•, 14] and require identification by molecular methods (see below). Both produce asexual hyaline septate hyphae from which cylindrical conidiogenous cells are produced (Table 2). Pale-brown ovoid conidia with truncate bases are typical, which are formed singly or in small clusters at the ends of the conidiophores, or from short annellidic necks arising directly from the hyphae (Figs. 1c and e).
The features of S. dehoogii and P. minutispora are summarized in Table 2.
Molecular Methods
Given that histopathological and culture morphology methods have limitations in providing definitive species identification of the agents of Scedosporium/Pseudallescheria infection, an increasing number of molecular techniques have been developed. The high resolving power of molecular methods has enhanced the laboratory’s capabilities to both rapidly identify species and to genetically characterize members of the Scedosporium/Pseudallescheria species complex. In particular, polymerase chain reaction (PCR)-based methods have grown in popularity for the rapid detection of these fungi from clinical specimens and to identify cultured isolates. These approaches include multiplex PCR, real-time PCR (RT-PCR), restriction fragment length polymorphism (RFLP) tests, isothermal methods, probe-based assays and DNA sequencing.
Multiplex Polymerase Chain Reaction (PCR)
Multiplex PCR involves the amplification by PCR of two or more loci simultaneously in one reaction using species-specific primers based on target sequence data. There are few reports with regards to the use of multiplex PCR to enable diagnosis of scedosporiosis.
Harun et al. [32] developed a multiplex system using species-specific primers targeting the fungal internal transcribed spacer (ITS1/2) regions and the 28S ribosomal DNA (rDNA) gene to detect S. aurantiacum, S. prolificans, and the P. boydii species complex/P. apiosperma directly from respiratory tract specimens. The multiplex assay showed 100 % specificity and sensitivity using known cultures of each of the three species or species groups and then underwent further validation using DNA extracts from sputum specimens from cystic fibrosis patients compared to culture on media selective for Scedosporium. Twenty-nine samples from 11 patients yielded growth of Scedosporium. PCR was positive for 18/29 (62 %) samples or for 7/11 (63.6 %) patients with a specificity of 97.2 %, positive predictive value of 78.3 % and negative predictive value of 94.1 % [32]. False negative results included six samples from a single patient. The reasons for this observation are uncertain. As for all multiplex PCR assays, competition between primers for the target sequences and reagents requires strict primer design and stechiometric control of the reaction components to avoid false positive and negative results [33]. Additional reasons for false negative results are insufficient target DNA and PCR inhibition. However, the sensitivity of an oligonucleotide array assay for the direct detection of fungi in sputum samples, also from cystic fibrosis samples, was much higher (100 %) [34•], which is most likely due to the different assay format (discussed below). Nonetheless, multiplex PCR is rapid and simple, lending itself as a suitable complement to culture for detecting Scedosporium from respiratory tract specimens where Scedosporium species may be of particular relevance, e.g. a lung transplant recipient with pulmonary infiltrates; however, further evaluation of multiplex assays, such as the one described above, is required to determine their position in routine care.
Oligonucleotide Array Assay
The principle of using oligonucleotide array formats to provide species identification of fungi is not new. Hsiao et al. [35] performed one of the larger evaluations following the development of an ITS1/ITS2-directed array to identify 64 species (32 genera) of clinically important fungi. Probes were PCR amplified using the primer sets ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′), and ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [36]. The sensitivity and specificity of the array was 98.3 % and 98.1 %, respectively. In a similar fashion, Bouchara et al. [34•] used an oligonucleotide array for the direct detection of fungi in sputum samples of cystic fibrosis patients. The array was constructed using species-specific probes, designed from the ITS1 or ITS2 regions of 20 species, including Aspergillus, Candida, and the Scedosporium/Pseudallescheria species complex. The array was validated with pure cultures, yielding a sensitivity of 100 % and specificity of 92.2 %. It was then used to directly detect fungi, including Scedosporium, in 57 sputum samples from 39 cystic fibrosis patients, and the results were compared with those obtained by culture. For 16 sputum samples, the results obtained by the array corresponded with those obtained by culture. For 33 samples, the array detected more fungal species than culture did, while the reverse was found for eight samples. Although not specific for Scedosporium species, such array formats are useful for the simultaneous detection of multiple fungal species that may be present in a clinical sample. The drawback of this approach is cost and the need for batch testing for efficiency.
Real time PCR
RT-PCR has the advantage of higher sensitivity and being capable of quantifying the number of molecules per PCR cycle. Most systems use fluorescently-labeled primers, probes, or dyes, and measure fluorescent signals from one or more reactions during each amplification cycle in real time. As for multiplex assays, experience with using RT-PCR to detect and identify Scedosporium species is limited.
Castelli et al. developed and validated two separate ITS-based RT-PCR assays for species-specific targeting of S. prolificans and S. apiospermum using clinical strains and then a murine model of invasive infection [37]. A total of 14 clinical strains and 141 samples, including blood, serum, and lung samples from infected CD1 mice, were analyzed. Results gave 100 % specificity, with a detection limit of 10 fg/μl DNA for both assays. Assay sensitivity for the S. prolificans-specific PCR assay was 100 % for cultured clinical strains, 95.5 % for lung tissues (n = 45), 85 % for serum (n = 18), and 83.3 % for blood (n = 20). For S. apiospermum-infected mice, the sensitivities were 100 % for clinical strains and 97.2 %, 81.8 %, and 54.5 % for lung tissues (n = 36), serum (n = 11), and blood (n = 11), respectively [37]. Although RT-PCR is simple and rapid with high analytical sensitivity and specificity, its use in routine clinical diagnostics for Scedosporium infections has not been applied. The advantage of rapid results, precise detection, and possible simultaneous quantification of pathogenic Scedosporium species justifies further testing in a diagnostic setting.
Pan-Fungal PCR
Multiple in-house pan-fungal PCR assays have been published and are beyond the scope of this review [38]. By amplifying the ITS regions (ITS1, ITS2, or both) for instance, followed by DNA sequencing of the amplicon, species identification can be achieved for Scedosporium species.
In one ITS1-directed pan-fungal PCR assay that included the detection of S. prolificans, the sensitivities were reported to be either 97 % or 68 % when applied on fresh tissue specimen or paraffin-embedded specimen, respectively [39]. Identification within members of the P. boydii complex may not be possible using only ITS-based PCR assays (see identification of cultures below).
DNA Sequencing
Nucleotide acid sequencing technology coupled together with greater computing power and bioinformatic analysis systems have increased the ability to resolve Scedosporium and Pseudallescheria species. The primary sequence targeted for nucleotide sequencing is the multiple-copy ITS regions of the rDNA gene.
Delhaes et al. [16•] sequenced the ITS1-5.8S-ITS2 regions, and phylogenetic analysis revealed three distinct clades—S. apiospermum/P. boydii species complex, with an intra-species sequence variation of 2.2 %, and S. prolificans and S. aurantiacum displaying no sequence variation [16•]. As such, a disadvantage of the ITS region is its insufficient variability to distinguish between species within complexes, and may not allow for a correct identification of later classified species within the S. apiospermum/P. boydii species complex. To increase the resolution between species, housekeeping genes such as β-tubulin (β-TUB), β-actin (β-ACT), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), calmodulin (CAL), and others have been utilized for sequence analysis. Gilgado and colleagues [13•, 14] sequenced regions of the ITS, CAL, and β-TUB genes to reclassify the P. boydii species complex, and revealed that P. boydii and S. apiospermum are two separate species [13•, 14, 15].
The continued reorganization of fungal species within the Scedosporium/Pseudallescheria species complex creates inconsistencies in the species name of sequences deposited in reference databases such as GenBank, European Molecular Biology Laboratory (EMBL) and DNA Databank of Japan (DDBJ), as the previously used names have not been changed in those databases after reclassification had occurred, leading to potential incorrect species identifications [40]. Within the International Society for Human and Animal Mycology (ISHAM) Working Group for DNA barcoding of medical fungi, a new quality-controlled ITS sequence database was developed; one of its aims is to resolve nomenclature inconsistencies. This database can be accessed at http://www.mycologylab.org.
Restriction Fragment Length Polymorphism
PCR-RFLP is a well-established technique for identification of fungi based on the digestion of h-PCR-amplified fragments with specific restriction endonuclease enzymes. The enzymatic digestion results in different banding patterns according to the fragment sizes specific for a certain species.
Delhaes et al. [16•] applied PCR-RFLP analysis to separate S. prolificans from S. apiospermum/P. boydii species complex and S. aurantiacum (n = 146 Scedosporium isolates). The ITS1-5.8S-ITS2 regions were PCR-amplified using SR6R (5′-AAGTARAAGTCGTAACAAGG-3′) and LR1 (5′-GGTTGGTTTCTTTTCCT-3′) primers and double digested with Sau96I and HhaI restriction endonucleases. Gel electrophoresis revealed one RFLP profile specific to S. prolificans and two RFLP profiles for isolates previously phenotypically identified as S. apiospermum, but now with one profile each corresponding to S. apiospermum/P. boydii and S. aurantiacum [16•].
Similarly, Lackner et al. [41] amplified the ITS regions with the primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) to differentiate most haplotypes of Pseudallescheria and Scedosporium species. This work employed the restriction enzymes AluI, HaeIII, HinfI, HindIII for the differentiation of S. dehoogii, S. prolificans, S. aurantiacum, P. apiosperma and P. boydii, or TauI for the differentiation of P. minutispora from P. boydii. However, for the identification of P. apiosperma/S. apiospermum and P.boydii, species-specific β-tubulin 2-targeted primers were needed. Pseudallescheria fusoidea remained difficult to distinguish from P. boydii. Whilst RFLP profiles are reproducible, to fully resolve all Scedosporium species a large number of enzymes are required. Depending on the local epidemiology of infection, one can expect that at least two enzymes are needed to detect the more common medically relevant species [16•]. This may not be time-effective for a busy laboratory.
Rolling Circle Amplification (RCA)
Rolling circle amplification (RCA) is a robust and simple technique for distinguishing closely-related species, demonstrating high specific detection of target nucleic sequences down to the single nucleotide level [42•]. RCA utilizes circularizing oligonucleotide (padlock) probes and specialized DNA polymerases under isothermal conditions to replicate short, single-stranded DNA circles [42•]. The padlock probes are designed using target sequences based on housekeeping genes such as the ITS region, and can be performed within 2 h isothermally using a water bath, heating block or thermocycler, and is typically performed in RT-PCR systems with signal detection within 15 min of initiation of the RCA reaction.
Two studies used ITS-directed RCA assays as a means of identifying Scedosporium species [43, 44]. Zhou et al. [43] clearly differentiated S. prolificans from S. apiospermum/P. boydii, whilst Lackner et al. [44] used RCA to distinguish at least seven to eight species within the Scedosporium/Pseudallescheria species complex. Both reports indicated that the technique was simple, rapid, and cost effective in comparison with DNA sequencing.
Proteomic Approaches
An alternative approach to the rapid identification of members of the Scedosporium/Pseudallescheria complex is to detect the proteins of intact fungal cells through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The principles of MS, its components and extraction methods for analysis of microorganisms have recently been reviewed [45–47]. In comparison with bacteria and yeasts, the identification of colonies of filamentous fungi by MALDI-TOF MS has proved more challenging. Firstly, standardization of growth conditions, form of fungal growth for analysis (e.g. conidia vs. hyphae), and protein extraction methods are necessary to achieve reproducible profiles [48, 49•, 50, 51]. At present, the construction of appropriate spectral databases and extension of those provided by manufacturers are required [49•]. Furthermore, melanin pigment in many moulds can inhibit ionization [52]. Nonetheless, MALDI-TOF MS has good potential to replace phenotypic and even gene sequencing methods for the identification of filamentous fungi as it is accurate, inexpensive, and results are available in <2 h.
Coulibaly et al. [53] analyzed 25 Scedosporium/Pseudallescheria strains (22 reference and three clinical isolates) by MALDI-TOF MS using α-cyano-4-hydroxycinnamic acid (CCA) matrix and employing the BioTyper software (Bruker Daltonics, Germany). Neither culture medium nor protein extraction methods (formic acid vs. trifluoroacetic acid) influenced the quality of MS identification; however, identification was optimal when performed on cultures between 3 and 5 days old. Spectra were reproducible after a month of subculture. A limitation of the study was the small number of clinical isolates studied [53]. In another study, the same researchers, using a standardized extraction procedure and by constructing their own spectral library, demonstrated that five of seven S. apiospermum strains were identified correctly, with the remaining two strains not being able to be identified [50].
More recently, Lau and colleagues demonstrated the importance of interrogating databases that contain spectra obtained under similar growth and extraction procedures. They developed a simple extraction method comprising mechanical lysis followed by modified formic acid extraction, for use on filamentous fungal cultures on solid media [49•]. Using this extraction procedure, high-quality, reproducible spectra were obtained which were unaffected by culture medium, colony age, or incubation conditions, including for Scedosporium species. All 11, 9 and 4 strains of S. apiospermum, P. boydii, and S. prolificans tested, respectively, were correctly identified to species level (many of these were reference strains). Against their in-house library, all 24 isolates were identified correctly; in contrast, none of the isolates tested were identified when interrogated against the Bruker library in use at the time of the study [49•].
The above results are encouraging; however, the evaluation of larger numbers of clinical isolates is essential to extend the current databases for increased discriminatory power and to determine the ability of MALDI-TOF MS to distinguish among species within the P. boydii species complex. In addition, investigations to evaluate its potential as a typing tool, including in tracking nosocomial transmission, such as is seen for other fungi (Aspergillus flavus and Candida parapsilosis) [54, 55], are warranted.
Molecular Typing of Scedosporium/Pseudallescheria Species
To understand the molecular epidemiology and trace possible outbreaks/nosocomial outbreaks, a variety of typing techniques have been developed and applied to some of the Scedosporium/Pseudallescheria species. The main methods include PCR fingerprinting [16•], random amplification of polymorphic DNA [56], amplified fragment length polymorphism (AFLP) [16•], and multilocus sequence typing (MLST) analysis (Meyer et al. and Tintelnot et al., unpublished data). As MLST typing is especially highly reproducible and as such is applicable for multicenter studies, species-specific MLST typing schemes have been developed; for S. aurantiacum, comprising the following genetic loci: ACT, CAL, EF1α, RBP2, SOD2, and β-TUB (Meyer et al., unpublished data); and S. apiospermum and Pseudallescheria boydii, comprising the following genetic loci: ACT, CAL, RPB2, SOD2 and BT2 (Tintelnot et al., unpublished data). These databases can be accessed at http://mlst.mycologylab.org for allele and sequence type identification. As their main use is in epidemiological studies rather than for routine clinical species identification, they will not be discussed in detail herein. For further information see Ruiz-Diez et al. [56], Harun et al. [57], Delhaes et al. [16•], and the review by Cortez et al. [7•].
Conclusion
Clinicians rely substantially on the clinical mycology laboratory to assist with the diagnosis of Scedosporium/Pseudallescheria infections. Visualization of the organism, although non-specific, is helpful in establishing certainty of diagnosis. Culture-based methods have recently seen increased utility, with the use of selective media to increase the yield of Scedosporium from non-sterile sites such as the respiratory tract. Molecular methods complement the culture methods, of which direct detection by PCR-based methods deserve more study. DNA sequencing, RCA, and MALDI TOF MS lend themselves as options for species identification of cultured isolates. Systematic comparison of all these approaches is necessary to quantify their utility in diagnosis.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Park BJ, Pappas PG, Wannemuehler KA, et al. Invasive non-Aspergillus mould infections in transplant recipients, United States, 2001-2006. Emerg Infect Dis. 2011;17:1855–64.
Husain S, Alexander BD, Munoz P, et al. Opportunistic mycelial fungal infections in organ transplant recipients: emerging importance of non-Aspergillus mycelial fungi. Clin Infect Dis. 2003;37:221–9.
Castiglioni B, Sutton DA, Rinaldi MG, Fung J, Kusne S. Pseudallescheria boydii (anamorph Scedosporium apiospermum) infection in solid organ transplant recipients in a tertiary medical centre and review of the literature. Medicine. 2002;81:333–48.
Heath CH, Slavin MA, Sorrell TC, et al. Population-based surveillance for scedosporiosis in Australia: epidemiology, disease manifestations and emergence of Scedosporium aurantiacum infection. Clin Microbiol Infect. 2009;15:689–93.
Pfaller MA, Diekma DJ. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J Clin Microbiol. 2004;42:4419–31.
Neofytos D, Horn D, Anaissie E, et al. Epidemiology and outcome of invasive fungal infection in adult haematopoetic stem cell transplant recipients: analysis of multicentre Prospective Antifungal Therapy (PATH) alliance registry. Clin Infect Dis. 2009;48:265–73.
Cortez KJ, Roilides E, Quiroz-Telles F, et al. Infections caused by Scedosporium spp. Clin Microbiol Rev. 2008;21:157–97. Relatively recent and very comprehensive review about the mycological aspects and scope of Scedosporium infections, including clinical aspects and treatment.
Husain S, Muñoz P, Forrest G, et al. Infections due to Scedosporium apiospermum and Scedosporium prolificans in transplant recipients: clinical characteristics and impact on antifungal agent therapy on outcome. Clin Infect Dis. 2005;40:89–99.
Katragkou A, Dotis J, Kotsiou M, Tamiolaki M, Roilides E. Scedosporium apiospermum infection after near-drowning. Mycoses. 2007;50:412–21.
Sahi H, Avery RK, Minai OA, et al. Scedosporium apiospermum (Pseudallescheria boydii) infection in lung transplant recipients. J Heart Lung Transplant. 2007;26:350–6.
Musk M, Chambers D, Chin W, Murray R, Gabbay E. Successful treatment of disseminated Scedosporium infection in 2 lung transplant recipients: review of the literature and recommendations for management. J Heart Lung Transplant. 2006;25:1268–72.
Guarro J, Kantarcioglu AS, Horré R, et al. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med Mycol. 2006;44:295–327.
Gilgado F, Cano J, Gene J, Guarro J. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol. 2005;43:4930–42. One of the first indications that the concept of a single species of ‘Scedosporium apiospermum’ was incorrect. The work has paved the way for future studies of the complex taxonomy of Scedosporium/Pseudallescheria complex.
Gilgado F, Cano J, Gene J, Sutton DA, Guarro J. Molecular and phenotypic data supporting distinct species statuses for Scedosporium apiospermum and Pseudallescheria boydii and the proposed new species Scedosporium dehoogii. J Clin Microbiol. 2008;46:766–71.
Gilgado F, Gene J, Cano J, Guarro J. Heterothallism in Scedosporium apiospermum and description of its teleomorph Pseudallescheria apiosperma sp. nov. Med Mycol. 2010;48:122–8.
Delhaes L, Harun A, Chen SCA, et al. Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum. Emerg Infect Dis. 2008;14(2):282–90. One of the first descriptions of the epidemiological associations of the then novel species Scedosporium aurantiacum which was uncovered by the molecular identification technique of restriction fragment length polymorphism of the ITS region.
Gilgado F, Serena C, Cano J, Gene J, Guarro J. Antifungal susceptibilities of the species of the Pseudallescheria boydii complex. Antimicrob Agents Chemother. 2006;50:4211–3.
Harun A, Serena C, Gilgado F, Chen SC, Meyer W. Scedosporium aurantiacum is as virulent as S. prolificans, and shows strain-specific virulence differences, in a mouse model. Med Mycol. 2010;48 Suppl 1:S45–51.
De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/ Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–21.
Kimura M, Maenishi O, Ito H, Ohkusu K. Unique histological characteristics of Scedosporium that could aid in its identification. Pathol Int. 2010;60:131–6.
Walts AE. Pseudallescheria: an under diagnosed fungus? Diagn Cytopathol. 2001;25:153–7.
Collignon PJ, Macleod C, Packham DR. Miconazole therapy in Pseudallescheria boydii infection. Australas J Dermatol. 1985;26:129–32.
Albernaz V, Huston B, Castillo M, Mukherji S, Bouldin TW. Pseudallescheria boydii infection of the brain: imaging with pathologic confirmation. Am J Neuroradiol. 1996;17:589–92.
Dubeau F, Roy LE, Allard J, et al. Brain abscess due to Petriellidium boydii. Can J Neurol Sci. 1984;11:395–8.
Hayden RT, Isotalo PA, Parrett T, et al. In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn Mol Pathol. 2003;12:21–6.
Pihet M, Carrere J, Cimon B, et al. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis: a review. Med Mycol. 2009;47:387–97.
Rainer J, Kaltseis J, de Hoog SG, Summerbell RC. Efficacy of a selective isolation procedure for members of the Pseudallescheria boydii complex. Antonie Van Leeuwenhoek. 2008;93:315–22.
Kaltseis J, Rainer J, De Hoog GS. Ecology of Pseudallescheria and Scedosporium species in human-dominated and natural environments and their distribution in clinical samples. Med Mycol. 2009;47:398–405.
Henson OE. Dichloran as an inhibitor of mold spreading in fungal plating media: effects on colony diameter and enumeration. Appl Environ Microbiol. 1981;42:656–60.
Blyth CC, Harun A, Middleton PG, et al. Detection of occult Scedosporium species in respiratory tract specimens from patients with cystic fibrosis by use of selective media. J Clin Microbiol. 2010;48:314–6. Study comparing the utility of different media in the isolation of Sceodsporium species from the sputa of cystic fibrosis patients, and establishing the occult presence of these fungi in the absence of using selective media.
King Jr AD, Hocking AD, Pitt JI. Dichloran-rose bengal medium for enumeration and isolation of molds from foods. Appl Environ Microbiol. 1979;37(5):959–64.
Harun A, Blyth CC, Gilgado F, Middleton P, Chen SC, Meyer W. Development and validation of a multiplex PCR for detection of Scedosporium spp. in respiratory tract specimens from patients with cystic fibrosis. J Clin Microbiol. 2011;49:1508–12.
Henegariu O, Heerema NA, Dlouhy SR, Vance GH, Vogt PH. Multiplex-PCR: critical parameters and step-by-step protocol. Biogeosciences. 1997;23:504–11.
Bouchara JP, Hsieh HY, Croquefer S, et al. Development of an oligonucleotide assay for direct detection of fungi in sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2009;47:142–52. Oligonucleotide array development and demonstration of feasibility of use in detecting fungi directly from the sputa of cystic fibrosis patients.
Hsiao CR, Huang L, Bouchara JP, Barton R, Li HC, Chang TC. Identification of medically important molds by an oligonucleotide array. J Clin Microbiol. 2005;43:3760–8.
White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc; 1990. p. 315–22.
Castelli MV, Buitrago MJ, Bernal-Martinez L, Gomez-Lopez A, Rodriguez-Tudela JL, Cuenca-Estrella M. Development and validation of a quantitative PCR assay for diagnosis of scedosporiosis. J Clin Microbiol. 2008;46:3412–6.
Lau A, Chen S, Sleiman S, Sorrell T. Current status and future perspectives on molecular and serological methods in diagnostic mycology. Future Microbiol. 2009;4:1185–222.
Lau A, Chen S, Sorrell T, et al. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimen. J Clin Microbiol. 2007;45:380–5.
Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Koljalg U. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1:e59.
Lackner M, Klaassen CH, Meis JF, van den Ende AH, de Hoog GS. Molecular identification tools for sibling species of Scedosporium and Pseudallescheria. Med Mycol. 2012;50:497–508.
Demidov VV. Rolling-circle amplification in DNA diagnostics: the power of simplicity. Expert Rev Mol Diagn. 2002;2:542–8. One of the earlier, but clear and useful description of the principle and applications of rolling circle amplification in detecting nucleotide polymorphisms in microbiology and other areas.
Zhou X, Kong F, Sorrell TC, Wang H, Duan Y, Chen SC. Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rolling-circle amplification. J Clin Microbiol. 2008;46:2423–7.
Lackner M, Najafzadeh MJ, Sun J, Lu Q, de Hoog GS. Rapid identification of Pseudallescheria and Scedosporium strains by using rolling circle amplification. Appl Environ Microbiol. 2012;78:126–33.
Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603.
Kok J, Chen SC, Dwyer DE, Iredell JR. Current status of matrix-assisted laser desorption ionization-time of flight mass spectrometry in the clinical microbiology laboratory. Pathology. 2013;45(1):4–17.
Steensels D, Verhagen J, Lagrou K. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin Belg. 2011;66:267–73.
Santos C, Paterson RRM, Venancio A, Lima N. Filamentous fungal characterization by matrix-assisted laser desorption/ionization time of flight mass spectrometry. J Appl Microbiol. 2010;108:378–85.
Lau AF, Drake SF, Calhoun LB, Henderson CM, Zelany AM. Development of a clinically comprehensive database and a simple procedure for identification of moulds from solid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:1828–34. This article outlines a simple method employing cultures grown on solid media for use in MALDI-TOF MS analysis of filamentous fungi. It also establishes a database that supplements and complements commercial databases that has enhanced the ability to accurately and rapidly identify mould pathogens.
Cassagne C, Ranque S, Normand AC, et al. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2011;6:e28425.
Bader O. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics. 2013;13:788–99.
Buskirk AD, Hettick JM, Chipinda I, et al. Fungal pigments inhibit the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of darkly pigmented fungi. Anal Biochem. 2011;411:122–8.
Coulibaly O, Marinach-Patrice C, Cassgne C, Piarroux R, Mazier D, Ranque S. Pseudallescheria/Scedosporium complex species identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Med Mycol. 2011;49:621–6.
Chang PK, Ehrlich KC, Hua SS. Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. Int J Food Microbiol. 2006;108:172–7.
Pulcrano G, Roscetto E, Iula VD, Panellis D, Rossano F, Catania MR. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur J Clin Microbiol Infect Dis. 2012;31:2919–28.
Ruiz-Díez B, Martín-Díez F, Rodríguez-Tudela JL, Alvárez M, Martínez-Suárez JV. Use of random amplification of polymorphic DNA (RAPD) and PCR-fingerprinting for genotyping a Scedosporium prolificans (inflatum) outbreak in four leukemic patients. Curr Microbiol. 1997;35:186–90.
Harun A, Perdomo H, Gilgado F, et al. Genotyping of Scedosporium species: a review of molecular approaches. Med Mycol. 2009;47(4):406–14.
Acknowledgments
We thank Professor Josep Guarro, University of Reus, Spain, for his helpful critique regarding information in the tables in this article.
Compliance with Ethics Guidelines
ᅟ
Conflicts of Interest
Sharon Chen has received honoraria, travel expenses, and research grants from Pfizer, Merck, and Gilead. Marc Ramsperger has declared no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramsperger, M., Duan, S., Sorrell, T.C. et al. The Genus Scedosporium and Pseudallescheria: Current Challenges in Laboratory Diagnosis. Curr Clin Micro Rpt 1, 27–36 (2014). https://doi.org/10.1007/s40588-014-0001-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40588-014-0001-z