Abstract
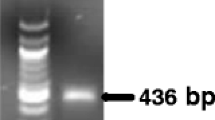
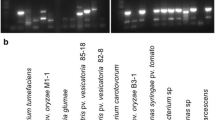
A novel real-time PCR assay was developed for the detection of Piggotia coryli, the causal agent of hazelnut anthracnose. A molecular tool for sensitive detection and quantification of P. coryli in symptomatic and asymptomatic host tissues was required to better understand P. coryli biology and epidemiology. Species-specific primers for P. coryli were designed based on the sequence of the β-tubulin gene. The amplification efficiency of pure target DNA was 93.2%, wet lab-tested limit of detection (LOD) was fixed at 8 pg/PCR reaction with maximum values of repeatability and reproducibility. In samples collected from infected orchards, the assay consistently revealed the presence of the pathogen in all symptomatic specimens, as well as in some asymptomatic tissues. This molecular tool will be of substantial aid in detecting and quantifying P. coryli, even in latent state and in different hazelnut tissues.



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding upon request.
References
Anderson JP, Gleason CA, Foley RC, Thrall PH, Burdon JB, Singh KB (2010) Plants versus pathogens: an evolutionary arms race. Funct Plant Biol 37(6):499–512. https://doi.org/10.1071/FP09304. PMID: 21743794; PMCID: PMC3131095
Arbefeville S, Harris A, Ferrieri P (2017) Comparison of sequencing the D2 region of the large subunit ribosomal RNA gene (MicroSEQ®) versus the internal transcribed spacer (ITS) regions using two public databases for identification of common and uncommon clinically relevant fungal species. J Microbiol Methods 140:40–46
Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morisset D (2014) Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol 37(2):115–126. https://doi.org/10.1016/j.tifs.2014.03.008
Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Calderon C, Ward E, Freeman J, McCartney A (2002) Detection of airborne fungal spores sampled by rotating-arm and Hirst-type spore traps using polymerase chain reaction assays. J Aerosol Sci 33:283–296
Druzhinina IS, Kopchinskiy AG, Kubicek CP (2006) The first 100 Trichoderma species characterized by molecular data. Mycoscience 47:55
Einax E, Voigt K (2003) Oligonucleotide primers for the universal amplification of β-tubulin genes facilitate phylogenetic analyses in the regnum Fungi. Org Divers Evol 3:185–194. https://doi.org/10.1078/1439-6092-00069
El-Sayed A, Kamel M (2020) Climatic changes and their role in emergence and re-emergence of diseases. Environ Sci Pollut Res 27:22336–22352. https://doi.org/10.1007/s11356-020-08896-w
Falacy JS, Grove GG, Mahaffee WF et al (2007) Detection of Erysiphe necator in air samples using the polymerase chain reaction and species-specific primers. Phytopathology 97:1290–1297
Fraaije BA, Cools HJ, Fountaine J et al (2005) Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology 95:933–941
Gervasi F, Scortichini M (2009) Detection of Pseudomonas avellanae from hazelnut twigs by TaqMan real-time PCR. J Plant Pathol 573–578
Gianetti G, Cravero S, Morone C (1994) Further observations about bioepidemiology of Monostichella coryli (Deuteromycetes, Melanconiales), causal organism of bud-rot of hazel. Acta Horticulturae (Netherlands)
Hughes KJ, Tomlinson JA, Giltrap PM et al (2011) Development of a real-time PCR assay for detection of Phytophthora kernoviae and comparison of this method with a conventional culturing technique. Eur J Plant Pathol 131:695–703
ISTAT (2021) Available at: https://www.istat.it. Accessed 10 Oct 2021
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Luo Y, Lichtemberg PS, Niederholzer FJ et al (2019) Understanding the process of latent infection of canker-causing pathogens in stone fruit and nut crops in California. Plant Dis 103:2374–2384
Mancini V, Murolo S, Romanazzi G (2016) Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol 65:691–703. https://doi.org/10.1111/ppa.12515
Martinelli F, Scalenghe R, Davino S et al (2015) Advanced methods of plant disease detection. A review. Agron Sustain Dev 35:1–25. https://doi.org/10.1007/s13593-014-0246-1
Mazzaglia A, Drais MI, Turco S et al (2021) First report of Erysiphe corylacearum causing powdery mildew on Corylus avellana in Spain. New Dise Rep
Merkes CM, Klymus KE, Allison MJ et al (2019) Generic qPCR Limit of Detection (LOD)/Limit of Quantification (LOQ) calculator. R Script. US Geol Surv Code Release
Migliorini D, Ghelardini L, Tondini E et al (2015) The potential of symptomless potted plants for carrying invasive soilborne plant pathogens. Divers Distrib 21:1218–1229
Molnar TJ, Walsh E, Capik JM et al (2013) A real-time PCR assay for early detection of eastern filbert blight. Plant Dis 97:813–818
O’Donnell K, Ward TJ, Geiser DM et al (2004) Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol 41:600–623
Pesante A (1966) Disseccamento dei rametti di Nocciòlo causato da un Gloeosporium. Boll. Lab. sper. Oss Fitopatol NS 27:17–21
Pilotti M, Tizzani L, Brunetti A et al (2010) Molecular identification of Fomitiporia mediterranea on declining and decayed hazelnut. J Plant Pathol 115–129
Pothier JF, Pagani MC, Pelludat C et al (2011) A duplex-PCR method for species-and pathovar-level identification and detection of the quarantine plant pathogen Xanthomonas arboricola pv. pruni. J Microbiol Methods 86:16–24
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. https://doi.org/10.1385/1-59259-192-2:365. PMID: 10547847
Sabelis MW, Bruin J (1996) 1.5.3. Evolutionary ecology: Life history patterns, food plant choice and dispersal. In: Lindquist EE, Sabelis MW, Bruin J (eds) World Crop Pests, Elsevier, Volume 6, pp 329–366. ISSN: 1572-4379; ISBN: 9780444886286. https://doi.org/10.1016/S1572-4379(96)80020-0
Scortichini M, Marchesi U (2001) Sensitive and specific detection of Pseudomonas avellanae using primers based on 16S rRNA gene sequences. J Phytopathol 149:527–532
Turco S, Bastianelli G, Morales-Rodrìguez C et al (2021) Development of a TaqMan qPCR assay for the detection and quantification of Gnomoniopsis castaneae in chestnut tissues. For Pathol e12701
Valasek MA, Repa JJ (2005) The power of real-time PCR. Adv Physiol Educ 29:151–159
West JS, Kimber RBE (2015) Innovations in air sampling to detect plant pathogens. Ann Appl Biol 166:4–17
Ye J, Coulouris G, Zaretskaya I et al (2012) Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134. https://doi.org/10.1186/1471-2105-13-134
Funding
This work has been supported by the European Commission under the Grant Agreement number 774571 (project PANTHEON ‘Precision farming of hazelnut orchards’).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
42161_2023_1326_MOESM1_ESM.docx
Supplementary file1 Melting curve analysis from the real time PCR assay, the peaks at 85.8 °C correspond to the dissociation curve of four specific amplification products obtained (DOCX 35 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Drais, M.I., Turco, S., D’Attilia, C. et al. Development of a quantitative PCR assay for the detection of Piggotia coryli, the causal agent of hazelnut anthracnose. J Plant Pathol 105, 507–516 (2023). https://doi.org/10.1007/s42161-023-01326-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-023-01326-z