Patterns of Fungal Community Assembly Across Two Culex Mosquito Species
- Division of Biology, Kansas State University, Manhattan, KS, United States
In the aquatic environment, mosquito larvae encounter bacteria and fungi that assemble into bacterial and fungal communities. The composition and impact of mosquito-associated bacterial community has been reported across larvae of various mosquito species. However, knowledge on the composition of mosquito-associated fungal communities and the drivers of their assembly remain largely unclear, particularly across mosquito species. In this study, we used high throughput sequencing of the fungal Internal transcribed spacer 2 (ITS2) metabarcode marker to identify fungal operational taxonomic units (OTUs) and amplicon sequence variants (ASVs) associated with field-collected Culex restuans and Culex pipiens larvae and their breeding water. Our analyses identified diverse fungal communities across larval breeding sites collected on a fine geographic scale. Our data show that the larval breeding site is the major determinant of fungal community assembly in these mosquito species. We also identified distinct fungal communities in guts and carcasses within each species. However, these tissue-specific patterns were less evident in Cx. restuans than in Cx. pipiens larvae. The broad ecological patterns of fungal community assembly in mosquito larvae did not vary between OTU and ASV analyses. Together, this study provides the first insight into the fungal community composition and diversity in field collected Cx. restuans and Cx. pipiens larvae using OTUs and ASVs. While these findings largely recapitulate our previous analyses in Aedes albopictus larvae, we report minor differences in tissue-specific fungal community assembly in Cx. restuans larvae. Our results suggest that while the fungal community assembly in mosquito larvae may be generalized across mosquito species, variation in larval feeding behavior may impact fungal community assembly in the guts of mosquito larvae.
Introduction
Bacterial communities “i.e., microbiota” associated with mosquito larvae have been well-characterized across several mosquito species using culture-dependent and culture-independent methods (Rani et al., 2009; Chavshin et al., 2012; Gimonneau et al., 2014; Kim et al., 2015; Coon et al., 2016; Bascuñán et al., 2018; Galeano-Castañeda et al., 2019; Scolari et al., 2021). In addition, the impact of these communities on mosquito life history and vector competence have been reported (Gaio et al., 2011; Dennison et al., 2014; Coon et al., 2016, 2017; Wang et al., 2018). Knowledge on mosquito-fungus interactions is largely confined to fungal entomopathogens of mosquito larvae and adults (Christophers, 1952; Jenkins, 1964; Scholte et al., 2004; Tawidian et al., 2019) or yeasts as food source for mosquito larvae and support larval development to adulthood (Asahina, 1964; Timmermann and Briegel, 1996; Steyn et al., 2016; Coon et al., 2017; Souza et al., 2019). However, mosquito-fungus encounters are not restricted to entomopathogenicity but can include symbiotic interactions that may further impact mosquito life history. More recently, studies have characterized the fungal communities of several species of field collected mosquito adults, including Aedes aegypti, Aedes albopictus, Aedes japonicus, Aedes triseriatus, Anopheles coluzzii, and Culex quinquefasciatus (Muturi et al., 2016; Krajacich et al., 2018; Thongsripong et al., 2018; Luis et al., 2019). In addition, recent studies report the fungal community composition and assembly in field collected Ae. albopictus and Ae. aegypti larvae (Tawidian et al., 2021; Zouache et al., 2022). However, it remains unclear whether similar patterns in fungal community assembly are observed in the larvae of other mosquito species.
Mosquito larval breeding sites harbor rich bacterial and fungal communities that vary across large and small geographic scales (Gimonneau et al., 2014; Coon et al., 2016; Bascuñán et al., 2018; Shelomi, 2019; Tawidian et al., 2021). The microbiota associated with field-collected mosquito larvae is largely influenced by the bacterial community in the aquatic habitat they reside in. Indeed, studies report a considerable overlap of bacterial taxa among mosquito larvae and their breeding environment (Gimonneau et al., 2014; Kim et al., 2015; Coon et al., 2016; Scolari et al., 2021). Similarly, fungal communities associated with field-collected Ae. albopictus and Ae. aegypti larvae reflect those in the larval breeding site (Tawidian et al., 2021; Zouache et al., 2022). In addition, tissue-specific fungal communities were observed in Ae. albopictus larval guts and carcasses, driven by mosquito feeding behavior and fungal mode of nutrition (ecological guild) (Tawidian et al., 2021). To date, no studies have characterized the fungal communities in larvae of any other mosquito species or the aquatic habitat they inhabit.
The overall goal of this study was to determine whether the fungal community assembly observed in the larvae of Aedes mosquitoes also hold true for Culex restuans and Culex pipiens mosquito larvae. In the United States, Cx. restuans and Cx. pipiens are mosquitoes of public health concern due to the transmission of the causative agent of West Nile Virus by the adult females (Andreadis et al., 2004; Andreadis, 2012; Rochlin et al., 2019). Cx. restuans and Cx. pipiens larvae exhibit a temporal pattern of prevalence in urban and suburban areas in east-central United States with Cx. restuans larvae being abundant in early summer, whereas Cx. pipiens larvae dominate in mid- to late summer (Jackson and Paulson, 2006; Gardner et al., 2013; Johnson et al., 2015). Larvae of both species occupy natural and human-made breeding sites (e.g., woodland pools, tires, catch basins, and storm drains) (Yee, 2008; Rochlin et al., 2019). In this study, we collected Cx. restuans and Cx. pipiens L4 larvae from various natural and human made mosquito breeding sites in the midwestern U.S. city of Manhattan in Kansas. To determine the fungal community composition and diversity in breeding water and mosquito larval guts and carcasses, we amplified and sequenced the fungal internal transcribed spacer 2 (ITS2) metabarcode marker and analyzed the data using OTU and ASV-based approaches. Both approaches congruently identified the larval breeding habitat as the major driver of fungal community assembly in these mosquito species. In addition, we identified different fungal communities across mosquito tissues, likely driven by mosquito feeding behavior and fungal nutritional mode. However, the variation in fungal communities in the larval guts and carcasses was less evident in Cx. restuans larvae, potentially because of lesser larval feeding as reflected by the low OTU/ASV richness across Cx. restuans.
Materials and Methods
Sampling
A total of ten Cx. restuans and Cx. pipiens L4 mosquito larvae and a single water sample (50 mL) were collected from each of eight larval breeding sites in Manhattan, KS. One breeding site (site 1) was sampled during 2017, whereas the remaining seven sites were sampled during 2018. Breeding sites 2, 4, 5, 6, and 8 were man-made containers consisting of tires, plastic containers, and a mosquito oviposition cup lined with seed germination paper (Anchor Paper Co., St Paul, MN, United States) (Figure 1). Sites 1, 3, and 7 were natural mosquito breeding sites consisting of small ponds (Figure 1). Site 1 was revisited a year after the first collection, and the revisited collection is referred to as site 3. Site 4 was revisited 2 months after the first collection and is referred to as site 5. Table 1 depicts sampling locations, dates, and mosquito species found at each larval breeding site. Mosquito larvae were collected in their breeding environments and transported to the laboratory in plastic containers (Bare Eco-Forward Rpet Deli Container).
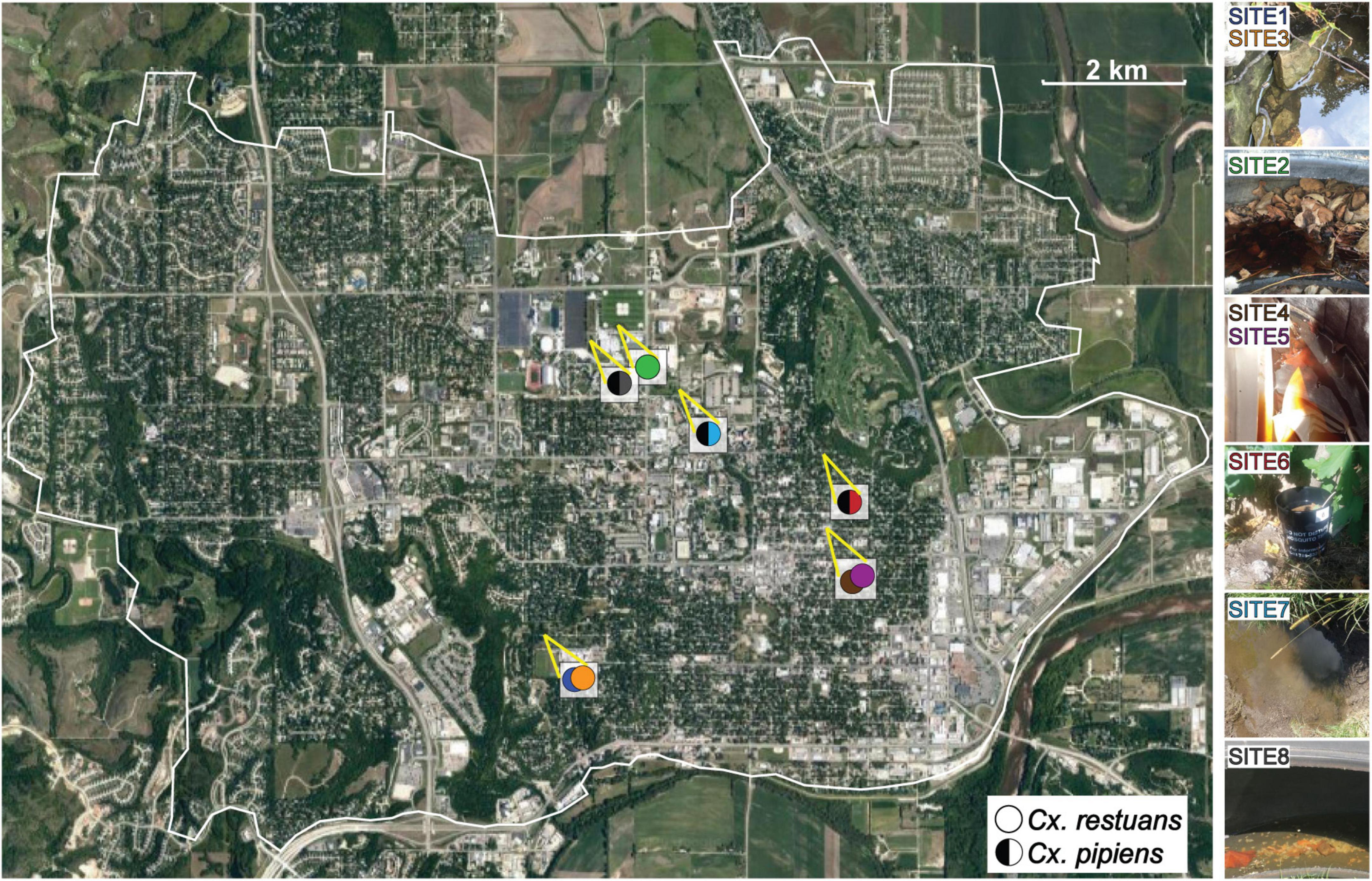
Figure 1. Map depicting the collection sites of Cx. restuans and Cx. pipiens mosquito larvae and their breeding water in Manhattan, KS. Solid circles indicate Cx. restuans breeding sites while circles with half left black symbol indicate Cx. pipiens breeding sites. Cx. restuans breeding sites 1 and 4 were revisited in a May (site 3) and July (site 5) 2018, respectively.
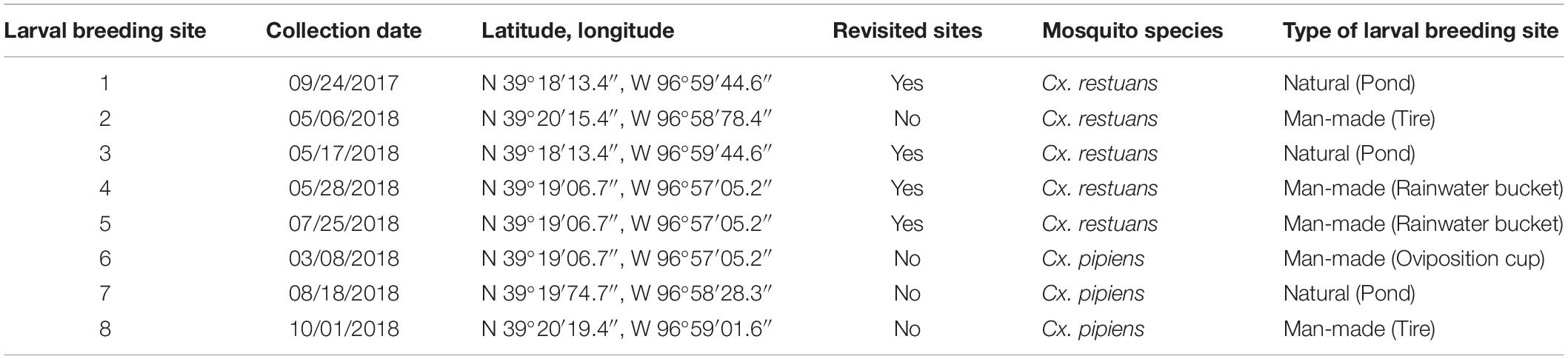
Table 1. Locations, dates, and types of larval breeding sites across Manhattan, KS during 2017 and 2018.
Sample Processing
In 2017, the larvae were incubated in their breeding water for 24 h at 27°C with 75% RH prior to gut dissection. In 2018, larvae were dissected immediately. Mosquito dissections were performed as described previously (Tawidian et al., 2021). In brief, larvae were washed six times with sterile Milli-Q water prior to gut dissection to eliminate fungal carryover from the breeding water and decapitated to remove loosely attached fungi from the breeding water on the mosquito mouth brushes. Sterile forceps and dissecting pins were used to dissect the larval guts, which included the gastric caeca, anterior midgut, posterior midgut, Malpighian tubules, and hindgut from the remainder of the body (carcass). Sterile net pieces were used as dissection controls to identify fungi introduced during the dissection process and to be subsequently removed from the community data. The larval breeding waters, dissected mosquito guts and carcasses, and controls were stored at −80°C until further processing.
The larval breeding water was filtered through 1-micron nuclepore membranes (Whatman®) immediately prior to nucleic acid extraction. From each of the collection sites, nucleic acids were extracted from ten individual guts and ten corresponding carcasses as well as a water sample and dissection control using the DNeasy® PowerSoil® Kit (MoBio Laboratory, Carlsbad, CA, United States) following the manufacturer’s instructions with minor modifications. The extracted DNA was quantitated and standardized to 2 ng/μL concentration using the NanoDropTM 2000/2000c spectrophotometers (Thermo Scientific, Waltham, MA, United States).
Total Nucleic Acid Extraction and Mosquito Identification
We identified mosquitoes by PCR amplification of the mosquito ITS2 region using the forward 5.8F (5′-TGTGAACTGCAGGACACATG-3′) and reverse 28R (5′-ATGCTTAAATTTAGGGGGTA-3′) primers following the protocol described by Collins and Paskewitz (1996). PCR cycling conditions were as follows: initial denaturation of 5 min at 94°C, 34 cycles of denaturing, annealing, and extension at 94°C for 30 s, 61°C for 30 s, 72°C for 30 s, respectively, followed by a final 72°C extension step for 5 min. The PCR included nuclease-free water as a PCR negative control. Amplicons were visualized by aliquoting 3 μL of each amplified product on 1% agarose gels in 1x TAE buffer at 100 V constant voltage for 30 min. Finally, the PCR products were purified on columns and sequenced using Sanger sequencing by Genewiz, Inc. (South Plainfield, NJ, United States).
Fungal Internal Transcribed Spacer Library Construction and Sequencing
The total nucleic acids from breeding water and mosquito tissues were used for PCR amplification of the fungal ITS2 region in triplicate PCRs with the barcoded forward fITS7 (5′-GTGARTCATCGAATCTTTG-3′) and reverse ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers, as described in Tawidian et al. (2021). PCR cycling conditions were as follows: an initial denaturation of 30 s at 98°C, 35 cycles of denaturing, annealing, and extension at 98°C for 10 s, 54°C for 30 s, 72°C for 1 min, followed by a final 72°C extension step for 9 min. The PCR included nuclease-free water as a PCR negative control. For positive control, we included a mock fungal community created with DNA from ten fungal species belonging to the phyla Ascomycota (Aspergillus niger, Chaetomium globosum, Penicillium notatum, Sordaria fimicola, Saccharomyces cerevisiae), Basidiomycota (Agaricus bisporus and Coprinus cinereus), Chytridiomycota (Phlyctochytrium acuminatum), and Mucoromycota (Phycomyces blakesleeanus and Rhizopus stolonifer) to determine the sequencing depth, error rate, and amplification with the fungal primers. Aliquots of 5 μL of each amplified product were visualized on 1% agarose gels in 1X TAE buffer at 100 V constant voltage for 30 min. The remaining volume from triplicate PCRs were pooled and purified using Mag-Bind® RXNPure plus (Omega Bio-Tek; Norcross, GA, United States). A total of 200 ng of amplicons from each sample were pooled for Illumina MiSeq sequencing. Illumina MiSeq adaptors were ligated onto the amplicon library using a NEBNext® DNA MasterMix for Illumina Kit (KAPA Biosystems, Wilmington, MA, United States), and sequences were generated using a MiSeq instrument (2 × 300 cycles: Illumina, San Diego, CA, United States) at the Kansas State University Integrated Genomics Facility (Manhattan, KS, United States).
Sequence Processing and Operational Taxonomic Unit Assignment
Fungal libraries were demultiplexed and the paired-end sequences processed through the mothur pipeline (v.1.38.1) (Schloss et al., 2009). As described in Tawidian et al. (2021), data denoising included filtering out ambiguous bases, mismatches to primers, and homopolymers longer than 10 bp. In addition, 110,151 chimeric sequences were removed using the VSEARCH algorithm (Rognes et al., 2016). The remaining 9,374,188 high-quality paired-end sequences were assigned to fungal taxa using naïve Bayesian classifier against the UNITE-curated International Nucleotide Sequence Database reference database (Wang et al., 2007; Abarenkov et al., 2010). Paired-end sequences assigned to Archaea, Plantae, and the protozoan phyla Cercozoa and Ciliophora were removed from the dataset through sequence processing in mothur. The filtered fungal sequences were pairwise aligned to generate a distance matrix that was clustered into fungal operational taxonomic units (OTUs) at a 97% similarity threshold using the average neighbor algorithm (UPGMA) using mothur (v.1.44.3) (Schloss et al., 2009). OTUs with less than ten sequence reads were considered as low abundance OTUs and removed from the dataset. In addition, OTUs assigned as unclassified fungi, unclassified Ascomycota, and unclassified Basidiomycota were manually cross-checked using NCBI’s Basic Local Alignment Search Tool (BLAST).1 BLAST revealed 296 non-fungal OTUs that were identified as algae (62.8%), plants (24.7%), protozoa (7.77%), insects (3.7%), fish (0.67%), and bacteria (0.33%). The non-fungal OTUs were removed from the dataset prior to downstream data analysis. Further, OTUs detected in the negative PCR and dissection controls (279 OTUs accounting for 529,196 reads) were also removed from the dataset. This resulted in a fungal dataset consisting of 1,670 OTUs and 695,922 reads for Cx. restuans samples and 1,543 OTUs, and 698,411 reads for Cx. pipiens samples (Supplementary Table 1).
The non-rarefied OTU dataset was further filtered prior to downstream data analysis to exclude all paired mosquito gut and carcass samples with fewer than 1,034 sequence reads. This eliminated all mosquito samples from two Cx. restuans breeding sites (sites 4 and 5), one Cx. pipiens breeding site (site 6), a single gut and carcass pair from site 2, two pairs of gut and carcass samples from site 1 and 7, respectively, and three pairs of gut and carcass samples from site 3. In addition, two Cx. restuans breeding sites (sites 2 and 5) were co-occupied by three and two Cx. pipiens mosquito larvae, respectively. The identified Cx. pipiens larvae were removed from downstream analyses because of the low sample size. The final dataset used in downstream analyses included five breeding water samples (three from Cx. restuans breeding sites and two from Cx. pipiens breeding sites), 21 guts and 21 carcasses of Cx. restuans larvae, and 18 guts and 18 carcasses of Cx. pipiens larvae. In addition, we also rarefied the data to the sequencing depth equal to the lowest yielding sample (1,034 sequence reads). Analysis of the rarefied data were congruent to our analysis of the non-rarefied data (data not shown). However, subsampling resulted in loss of 92% of the sequence reads, including the loss of the rare taxa in the samples.
Amplicon Sequence Variant Assignment
We used mothur (v.1.44.3) (Schloss et al., 2009) to compare the mosquito fungal community composition and assembly using OTUs and ASVs, while preserving the data denoising and cleanup performed in the OTU pipeline described above. For ASV assignment in mothur, pairwise aligned sequences were pre-clustered at a threshold of two nucleotide differences, prior to removing chimeric sequences through VSEARCH (Rognes et al., 2016). Upon removing chimeric sequences, ASVs were assigned to paired-end sequences using the UNITE-curated International Nucleotide Sequence Database reference database (Wang et al., 2007; Abarenkov et al., 2010). Similar to the OTU assignments, non-fungal ASVs and low abundance ASVs (accounting for less than ten reads) were removed from the dataset through the mothur pipeline. In addition, ASVs associated with the 296 non-fungal OTUs identified using BLAST were removed from the dataset. The final ASV dataset consisted of 3,303 ASVs, and 2,080,836 reads for Cx. restuans samples, and 2,548 ASVs, and 1,518,762 reads for Cx. pipiens samples (Supplementary Table 2). Similar to OTUs, ASVs were filtered to exclude all paired mosquito gut and carcass samples with fewer than 1,034 reads. The additional filtering steps eliminated the same mosquito samples and breeding sites as those listed with OTUs. Rarefaction analyses of the fungal ASVs were saturated for most samples (Supplementary Figure 1B) consistent with the high Good’s coverage (0.998 ± 0.003 SD), indicating that the sequencing depth was sufficient to capture the fungal diversity across most of the samples. Downstream ASV community analyses were performed as described for the OTUs. Results of the ASV community analyses are available in Supplementary Data Sheet 1.
Community and Statistical Analyses
To assess the fungal diversity and richness in breeding water and mosquito guts and carcasses, alpha diversity indices, including observed richness (SObs) and Shannon’s diversity (H’) were iteratively estimated (100 iterations) using mothur (v.1.44.3) (Schloss et al., 2009). The diversity indices were compared using non-parametric Wilcoxon signed-rank tests between paired mosquito guts and carcasses of each mosquito species separately. Breeding water samples were not included in the statistical analyses because of the low sample size after filtering. Wilcoxon signed-rank tests were performed using the GraphPad Prism version 8.4.3 for Windows (GraphPad Software, San Diego, CA, United States).
The Bray-Curtis dissimilarity matrices were computed for the fungal OTU data to compare and visualize the fungal community assemblages between the breeding water and mosquito guts and carcasses using non-metric multidimensional scaling (NMDS). To determine how much variation in the fungal community composition was inferred by larval breeding sites or mosquito tissue type, permutational multivariate analysis of variance (PERMANOVA) was performed. PERMANOVAs were performed using the vegan package in R.2 In addition, Wilcoxon signed-rank tests on the Bray-Curtis distances were used to assess the similarity of the fungal community composition in the breeding water to each mosquito tissue type. To identify the fungal OTUs that drive the fungal community variation across mosquito tissue types, we used the indicator value index (IndVal) analysis (Dufrêne and Legendre, 1997). In addition, the point-biserial correlation index (rpb) (Cáceres and Legendre, 2009) was used to determine the tissue niche preference of the OTUs associated with the mosquito guts and carcasses. Both analyses were performed using the indicspecies package in R with 9,999 permutations (see text footnote 2). Finally, to determine the ecological guild of the identified OTUs, we assigned them to ecological guilds as saprophytes, plant pathogens, endophytes, animal pathogens, lichen parasites, fungal parasites, epiphytes, and animal endosymbionts using the FUNGuild annotation tool (Nguyen et al., 2016).
Results
Temporal Prevalence of Culex Mosquitoes
We identified two Culex mosquito species collected from eight sites in Manhattan, KS, during 2017 and 2018 (Figure 1). Cx. restuans mosquito larvae were collected from five larval breeding sites, one in September 2017 and the remaining four during May-July 2018 (Table 1). While two Cx. restuans breeding sites were co-occupied by Cx. pipiens mosquito larvae, the prevalence of Cx. pipiens larvae was low in the early season. However, during August-October 2018, Cx. pipiens mosquitoes were more widespread and collected from three larval breeding sites (Table 1). These results show that both mosquito species are widely distributed and may have a largely complementary temporal pattern of prevalence in Manhattan, KS.
Fungal Community Composition in the Breeding Water and Mosquito Tissues
To determine the fungal community composition across larval breeding sites and mosquito tissues, we analyzed the fungal ITS2 metabarcode markers of breeding water as well as mosquito gut and carcass samples. Our analyses revealed 1,670 OTUs associated with Cx. restuans samples and 1,543 OTUs with Cx. pipiens samples. Rarefaction analyses indicated that the fungal OTUs were saturated for most samples (Supplementary Figure 1A). In addition, the average Good’s coverage across all samples was high (0.999 ± 0.001 SD). These results indicated that our sampling had sufficient sequencing depth and coverage to detect the fungal diversity across the samples.
To determine the fungal community composition across all samples, we plotted the relative abundances of the identified OTUs on the phylum and order levels (Figure 2). Twelve fungal phyla were identified accounting for 74.4% of total sequence reads. The phylum Ascomycota dominated with 45.8% of the sequences, followed by the phyla Basidiomycota (24.1%), Chytridiomycota (1.7%), Mucoromycota (1.7%), and Mortierellomycota (0.47%). A small proportion (0.63%) of the reads was assigned to seven rare phyla including Blastocladiomycota, Glomeromycota, Entorrhizomycota, Kickxellomycota, Monoblepharomycota, Rozellomycota, and Zoopagomycota (Figure 2). The remaining 25.6% of the reads were not classified beyond Fungi but were included in downstream analyses. In addition, the ascomycetous orders Pleosporales, Hypocreales, and Eurotiales were dominant and shared across most breeding water and mosquito samples (Figure 2). Within the phylum Basidiomycota, the order Ustilaginales was assigned the highest number of sequence reads (36.1%) followed by the order Agaricales (11.5%). As for the low abundance phyla, the majority of the remaining reads (90.5%) were assigned to the orders Mucorales and Mortierellales within the phylum Mucoromycota (Figure 2).
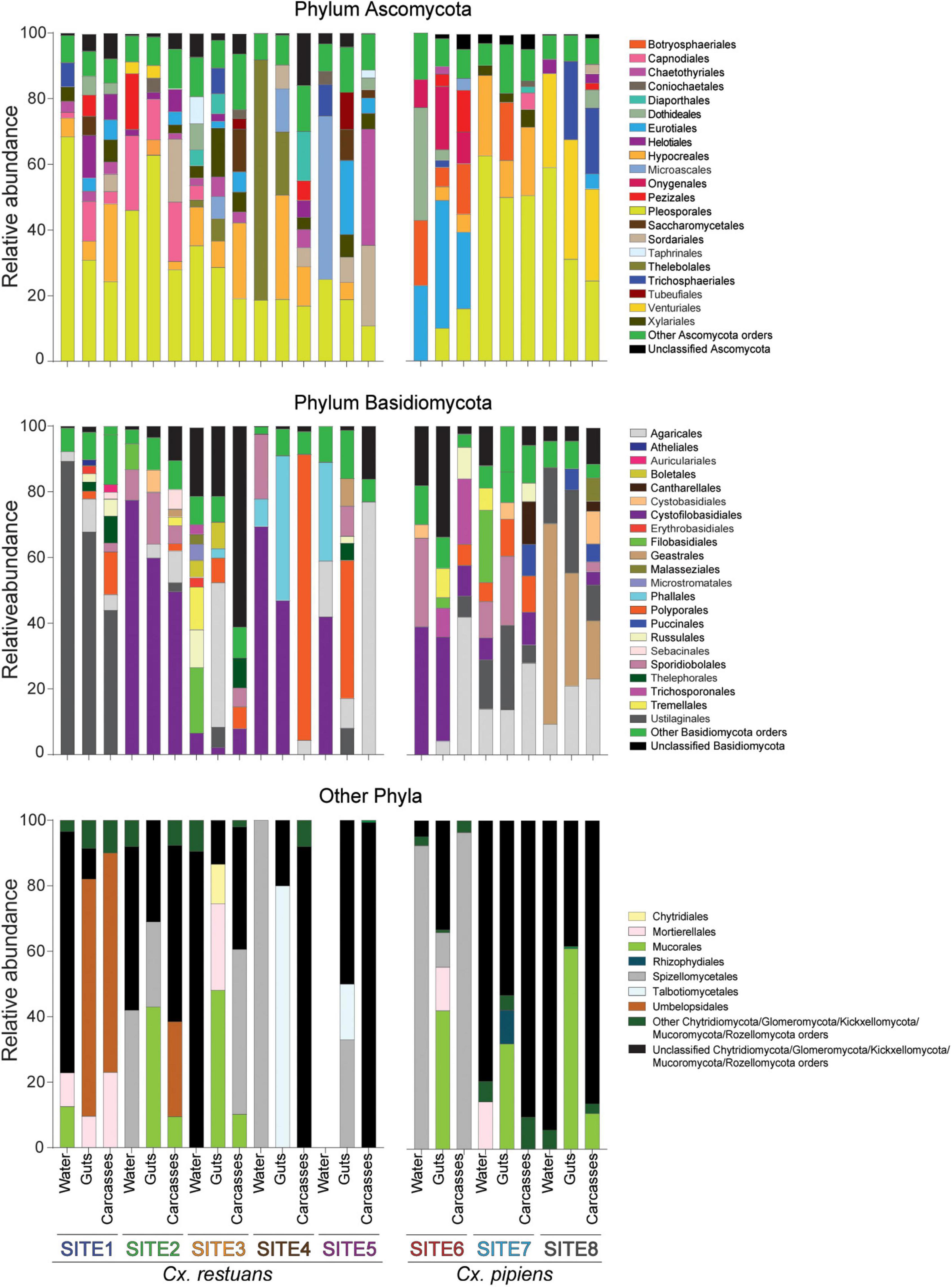
Figure 2. Bar plot of the relative abundances of fungal OTUs at the phylum and order levels across breeding water and larval guts and carcasses of both mosquito species. Gut and carcass samples presented here are the combined reads of all guts and carcasses within each larval breeding site. Low abundance orders within each phylum are combined into “other orders” category.
Fungal Community Composition in Culex Mosquito Larvae Is Driven by the Aquatic Habitat
To identify whether the larval breeding site drives fungal community assembly in Cx. restuans and Cx. pipiens mosquito larvae, we analyzed the Bray-Curtis community dissimilarities for each mosquito species separately. We visualized the community data using non-metric multidimensional scaling (NMDS) (Figure 3). We used permutational multivariate analysis (PERMANOVA) to determine whether fungal communities were distinct across Cx. restuans and Cx. pipiens breeding sites. These analyses revealed distinct fungal communities across larval breeding sites. In addition, PERMANOVA analysis showed significant differences in mosquito-associated fungal communities across larval breeding sites, suggesting that the aquatic breeding environment is a major driver of fungal community assembly in the larvae of both mosquito species (Figures 3A,B). In addition, fungal communities differed in composition across guts and carcasses of Cx. pipiens mosquito larvae with no evidence of tissue specificity in Cx. restuans larvae (Figures 3A,B).
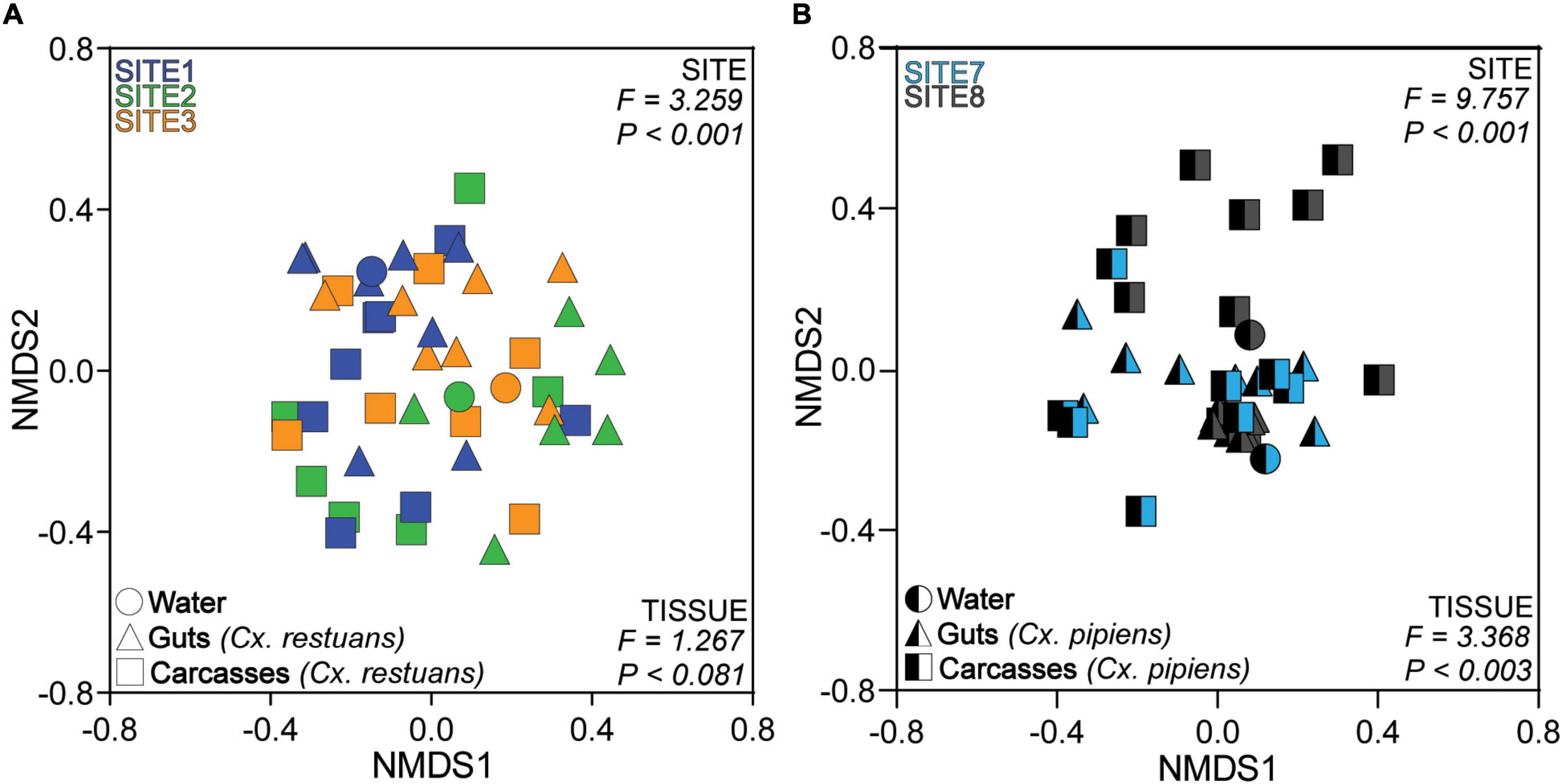
Figure 3. Non-metric Multidimensional Scaling (NMDS) ordination of the Bray-Curtis dissimilarity distances across larval breeding water and mosquito guts and carcasses. (A) NMDS of Bray-Curtis dissimilarity distances across Cx. restuans breeding water and guts and carcasses. (B) NMDS of Bray-Curtis dissimilarity distances across Cx. pipiens breeding water and guts and carcasses. The keys at the top left of each plot indicate the color designated to each larval breeding site, while the keys at the bottom left designate the shapes associated with each sample type. Samples cluster significantly by site as presented by the PERMANOVA results at the top right of each plot. Samples also cluster by tissue type as indicated by the PERMANOVA results at the bottom right of each plot.
Fungal Diversity Varies Across Mosquito Tissue Types
To identify whether fungal communities varied across mosquito tissues within each breeding site, we visualized the Bray-Curtis distances from individual guts and carcasses using NMDS (Supplementary Figure 2A) and compared them using PERMANOVAs. Our results revealed significant differences in fungal community composition among guts and carcasses for one of three Cx. restuans and two Cx. pipiens breeding sites. This was also evident when we plotted the paired Bray-Curtis dissimilarity distances from the breeding water to guts and breeding water to carcasses for each mosquito species across all breeding sites (Figure 4). The results revealed that larval guts were more similar to the breeding water than the paired carcasses, indicating that mosquito guts had fungal communities more similar to the breeding water than the carcasses did. While this pattern was evident for both mosquito species when analyzed across all breeding sites, within individual breeding sites, the fungal communities in Cx. restuans larval guts and carcasses did not significantly differ in their distance to the breeding water. This is likely because the distances between the guts and breeding water varied widely among sites in Cx. restuans larvae. This variability was not observed in the Cx. pipiens gut to breeding water distances (Supplementary Figure 2B).
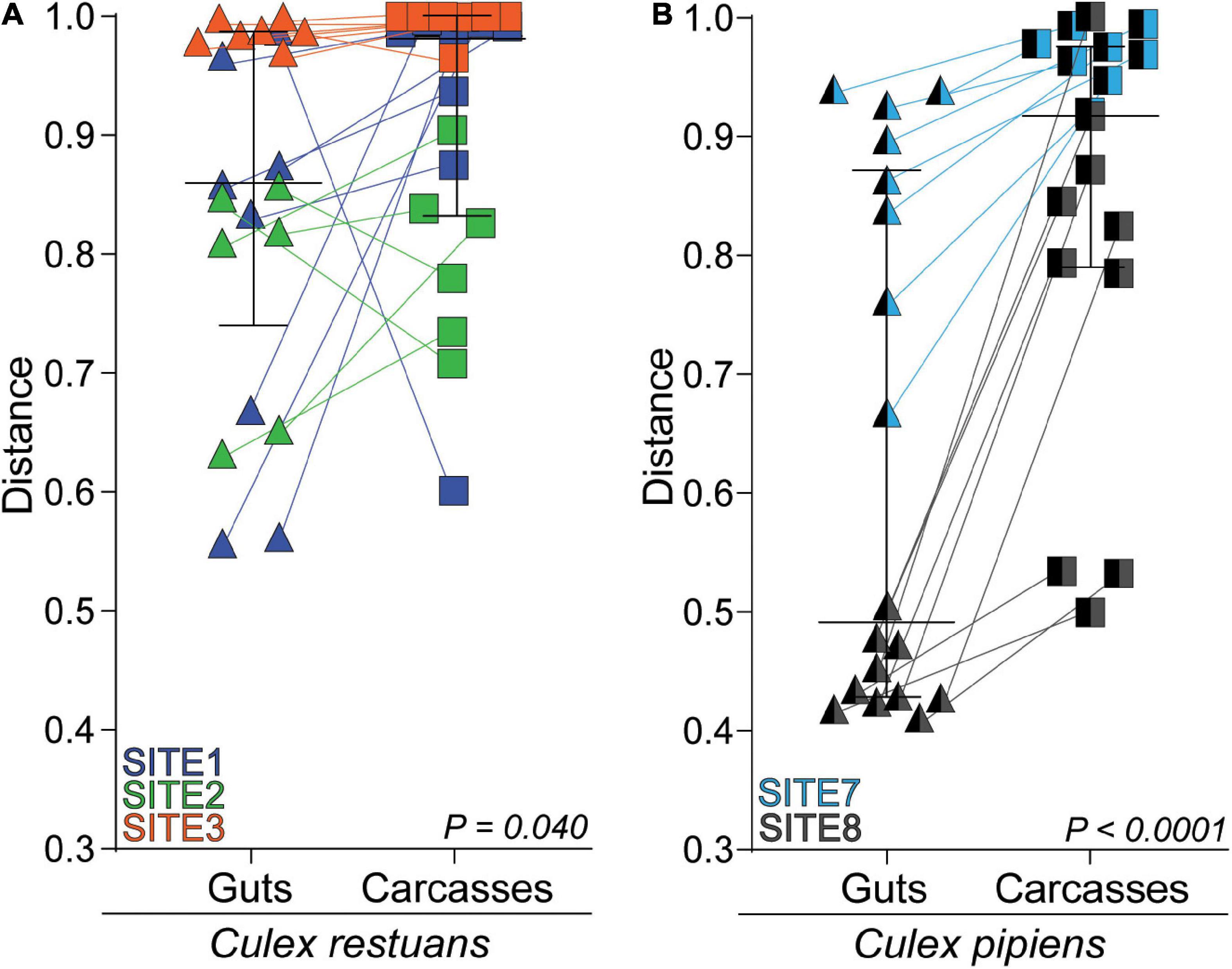
Figure 4. Bray-Curtis dissimilarity distances of fungal communities from the breeding water to larval guts vs. larval carcasses across all breeding sites. (A) Bray-Curtis distances for Cx. restuans guts and carcasses. (B) Bray-Curtis distances for Cx. pipiens guts and carcasses. Connecting lines shows matched individual mosquito gut and carcass samples. Statistical significance is inferred by a Wilcoxon signed-rank test.
In addition, our diversity analyses revealed variation in the fungal OTU richness (SObs) and diversity (Shannon’s H’) diversity across mosquito guts and carcasses (Figure 5). In Cx. restuans larvae, OTU richness was significantly higher in guts than in carcasses (Figure 5A), whereas Shannon’s diversity did not differ among mosquito tissues (Figure 5B). As for Cx. pipiens larvae, OTU richness as well as Shannon’s diversity were significantly higher in guts than carcasses (Figures 5C,D). While these results indicate differences in fungal community composition and diversity across guts and carcasses of both mosquito species, the observed tissue specificity was more evident in Cx. pipiens than in Cx. restuans larvae.
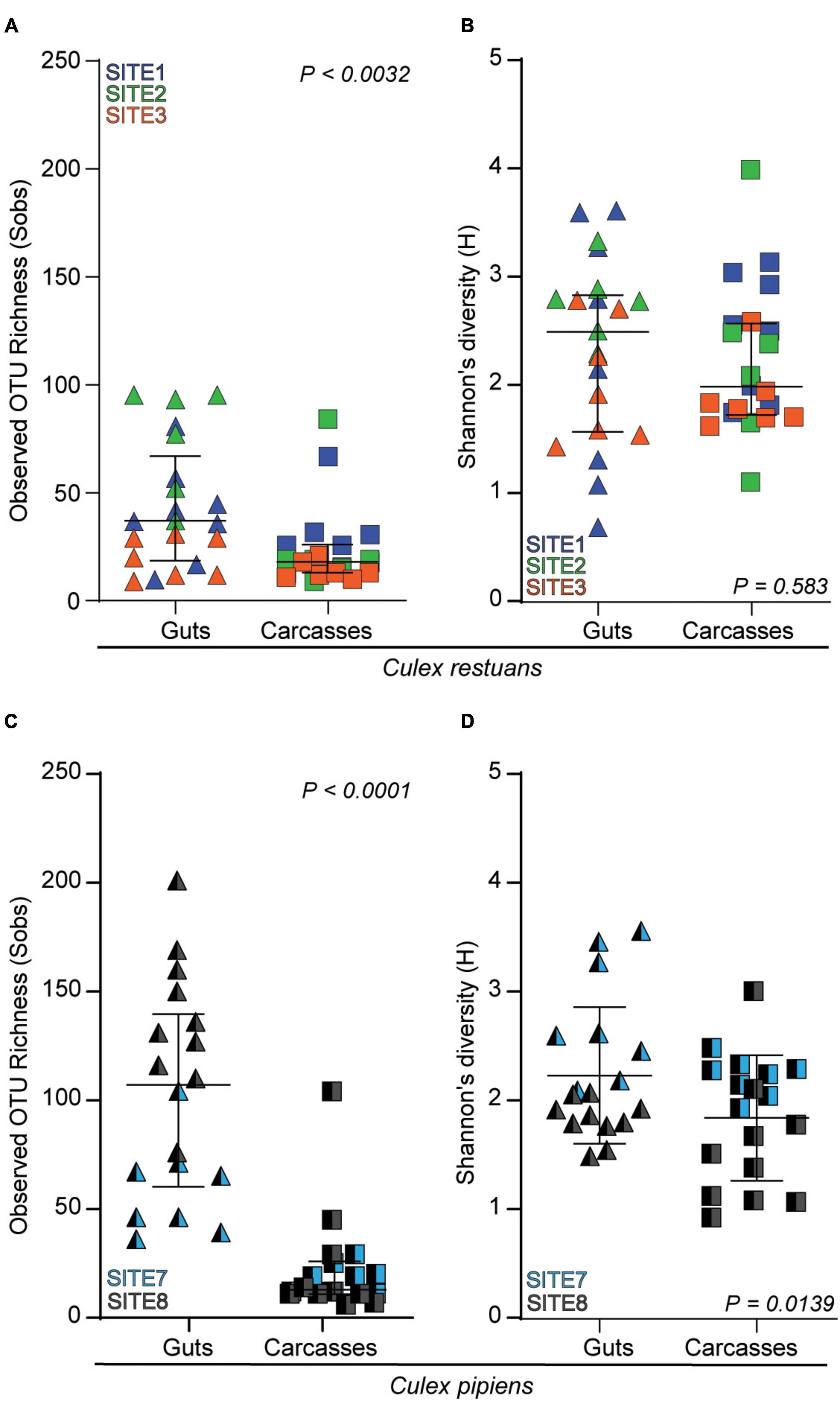
Figure 5. Alpha diversity indices as measured by observed OTU richness (Sobs) and Shannon’s diversity (H) of larval guts and carcasses across collection sites. (A) OTU richness in Cx. restuans guts and carcasses. (B) Shannon’s diversity in Cx. restuans guts and carcasses. (C) OTU richness in Cx. pipiens guts and carcasses. (D) Shannon’s diversity in Cx. pipiens guts and carcasses. Statistical significance was determined using a Wilcoxon signed-rank test.
Tissue-Specific Indicator Fungi Reflect Mosquito Feeding Behavior and Fungal Niche Preference
To identify the OTUs that drive tissue-specific patterns in fungal community assembly in Cx. restuans and Cx. pipiens mosquito larvae, we used indicator taxon analyses using the indicator value index (IndVal) (Figure 6 and Supplementary Table 3). We then used the FUNGuild annotation tool to assign the identified OTUs to ecological guilds. Three OTUs were identified as Cx. restuans gut indicators, whereas none were identified as indicators of carcasses (Figure 6A and Supplementary Table 3A). Two of three identified OTUs were assigned to unique ecological guilds (endophyte and fungal parasite). The third remaining OTU could not be assigned to an ecological guild. However, Cx. pipiens larval guts had higher number of indicators than Cx. restuans larvae. IndVal analysis identified 19 Cx. pipiens gut indicator OTUs (Figure 6B and Supplementary Table 3A), whereas carcasses were not assigned any indicators. The identified gut indicators OTUs were assigned to five ecological guilds [saprophyte (50%), plant pathogen (22.7%), endophyte (9.1%), animal pathogen (4.5%), and epiphyte (4.5%)] (Figure 6B and Supplementary Table 3A). The remaining 14.8% of indicator OTUs were not assigned to ecological guilds.
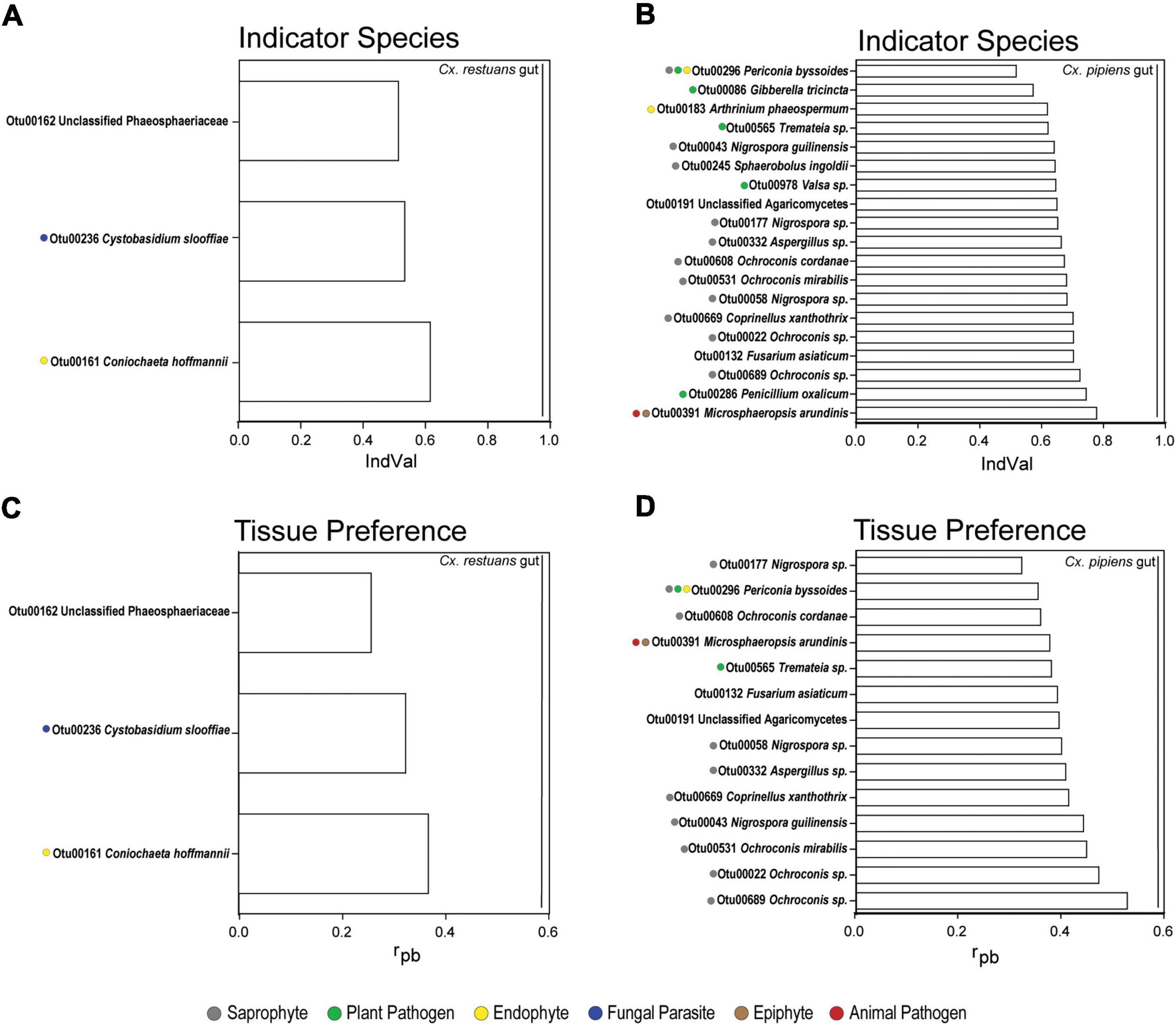
Figure 6. Indicator species analysis using indicator value index (IndVal) and point-biserial correlation index (rpb) of fungal OTUs identified in mosquito guts and carcasses across all breeding sites. (A) IndVal analysis of Cx. restuans gut indicator OTUs. (B) IndVal analysis of Cx. pipiens gut indicator OTUs. (C) OTU tissue preference across Cx. restuans guts and carcasses using rpb analysis. (D) OTU tissue preference across Cx. pipiens guts and carcasses using rpb analysis. Statistical significance is measured at P-value of 0.05 and with 9,999 permutations. Ecological guilds of each OTU identified using the FUNGuild annotation tool are presented by color coded solid circles.
Finally, we used the point-biserial correlation index (rpb) to determine the role of fungal niche preference on fungal community assembly across mosquito tissue types (Figure 6 and Supplementary Table 3B). Our results revealed three OTUs differentially abundant in Cx. restuans guts, all of which were also gut indicators (Figure 6C and Supplementary Table 3B). As for Cx. pipiens larvae, 14 OTUs were significantly associated with gut samples assigned to the ecological guilds [saprophyte (58.8%), plant pathogen (11.8%), endophyte (5.9%), animal pathogen (5.9%), and epiphyte (1.2%)] (Figure 6D and Supplementary Table 3D). The remaining 11.8% of OTUs were not assigned to ecological guilds. The rpb analyses did not identify any OTUs as significantly associated with the carcasses of either mosquito species.
Discussion
This study provides fundamental insight into the composition and diversity of fungal communities associated with Cx. restuans and Cx. pipiens mosquito larvae. In addition, we determine whether the observed fungal community assembly in mosquito larvae were similar across mosquito species. Overall, our OTU and ASV data demonstrate that the breeding site was the major determinant of fungal community assembly in the larvae of both mosquito species. In addition, we observed tissue-specific fungal community assembly in larval guts and carcasses. However, variation in fungal diversity and composition across mosquito tissues was more evident in Cx. pipiens larvae compared to Cx. restuans.
The fungal community associated with larval breeding water and mosquito guts and carcasses was dominated by the phyla Ascomycota and Basidiomycota, consistent with data reported in Ae. albopictus mosquito larvae collected in Manhattan, KS (Tawidian et al., 2021) and Aedes mosquito adults and larvae collected from the field (Muturi et al., 2016; Luis et al., 2019; Zouache et al., 2022). The observed phylum dominance was not surprising given the high prevalence of fungi in the phyla Ascomycota and Basidiomycota in habitats where mosquito larvae reside (Shelomi, 2019; Tawidian et al., 2021; Udujih Obinna Godwin et al., 2021; Zouache et al., 2022). In addition, it can also be attributed to the amplification bias of the ITS2 primers (fITS7 and ITS4) toward fungi belonging to these phyla with low affinity for early-diverging fungal lineages, e.g., including taxa in the Chytridiomycota (Bellemain et al., 2010; Blaalid et al., 2013; Bokulich and Mills, 2013).
The filamentous fungi and yeasts largely overlapped (42.6%) in Cx. restuans and Cx. pipiens mosquito larvae. The majority (96.4%) of fungal genera that overlapped across both Culex species were ascomycetes (68.7%) and basidiomycetes (27.7%). Similarly, an overlap of fungal genera (17.3%) was also observed between our Culex datasets and fungi detected by similar metabarcode sequencing of larval Ae. albopictus tissues (Tawidian et al., 2021). This overlap (95.9%) was also largely attributable to ascomycetes and basidiomycetes. In addition, a considerable proportion of fungi (62.9% genus level and 15.1% species level) isolated from field collected mosquito larvae, including Cx. pipiens were also identified in our dataset (Badran and Aly, 1995; Pereira et al., 2009; Steyn et al., 2016). The observed overlap of ascomycetes and basidiomycetes across larvae of different mosquito species can be explained by the ubiquity and co-occurrence of these taxa in freshwater ecosystems (Manohar and Raghukumar, 2013; Panzer et al., 2015; Lepère et al., 2019; Shelomi, 2019; Tawidian et al., 2021).
The overall goal of this study was to determine whether the drivers of fungal community assembly in mosquito larvae can be generalized across mosquito species. Our results show that fungal community assembly in Culex mosquito larvae is driven by the breeding site. This result corroborates our previous study assessing the fungal community assembly in Ae. albopictus larvae (Tawidian et al., 2021). Similarly, bacterial diversity in mosquito larvae also reflects that of breeding water, as reported for several species of field-collected mosquito larvae, including Ae. japonicus, Ae. triseriatus, Ae. aegypti, Ae. albopictus, Anopheles albimanus, An. coluzzii, Anopheles darlingi, Anopheles nuneztovari, Culex tharsalis, Cx. quinquefasciatus, and Cx. restuans (Duguma et al., 2013; Gimonneau et al., 2014; Kim et al., 2015; Coon et al., 2016; Bascuñán et al., 2018; Galeano-Castañeda et al., 2020; Scolari et al., 2021). To our knowledge, this is the first study that assesses the fungal community composition and diversity in field-collected Cx. restuans and Cx. pipiens mosquito larvae and their breeding water. We acknowledge that direct comparisons of fungal communities of Cx. restuans and Cx. pipiens larvae in our study are not possible, because the two species did not co-inhabit the larval breeding sites we sampled. However, we show that the fungal communities of both Cx. restuans and Cx. pipiens larvae cluster largely by site. Thus, these data provide support that the larval aquatic habitat is a major determinant of fungal community assembly in the larvae of both Culex species.
Our results further show variation in fungal community composition in the guts and carcasses of Culex mosquito larvae. While the fungal communities in the guts of Culex larvae were more similar to those in the breeding water than the carcasses, this pattern was more consistent in Cx. pipiens guts compared to Cx. restuans. The similarity of gut fungal community to that of the water can be attributed to the filter-feeding behavior of Culex larvae, which allows for the acquisition of fungi from the environment (Merritt et al., 1992). However, while similar, the gut fungal community of neither mosquito species was identical to that of the breeding water. This may be attributed to grazing behavior of Culex larvae on decaying plant detritus found in the aquatic habitat, in addition to the filter feeding behavior in the water column (Merritt et al., 1992; Yee et al., 2004; Winters and Yee, 2012). This notion is supported by the result of the Cx. pipiens indicator taxon analysis, in which the gut indicators were enriched with saprophytes, plant pathogens, and endophytes. These indicator taxa included obligate plant pathogens (family Ustilaginaceae or the rust diseases) and other plant pathogens, such as Gibberella tricincta (Desjardins, 2003; Geiser et al., 2014). These results corroborate earlier studies with Ae. albopictus larvae, in which the same taxa were indicators for the larval guts (Tawidian et al., 2021). Combined, these results suggest similar feeding behaviors of Cx. pipiens and Ae. albopictus larvae, and support earlier observations by Yee et al. (2004). Our indicator taxon analyses further showed that Cx. restuans guts were less enriched with plant-associated fungi and had lower OTU richness than Cx. pipiens guts, irrespective of the greater sample size. This suggests that Cx. restuans larvae fed less on plant detritus than Cx. pipiens larvae do, resulting in the observed differences in the gut fungal communities. These results corroborate reports on Cx. restuans feeding behavior (Winters and Yee, 2012), suggesting that Cx. restuans larvae affect detrital decay less than Ae. albopictus larvae.
In comparison, carcasses of both mosquito species had lower fungal diversity and richness than guts and breeding water. In contrast to the Ae. albopictus dataset (Tawidian et al., 2021), indicator species analysis did not reveal entomopathogenic/entomotoxigenic fungi associated with the carcasses of either Culex species. This is perhaps not surprising, given the low abundance of known entomopathogens in larval breeding environments. However, analysis of the ASV dataset identified Alternaria tenuissima (ASV1048) as significantly enriched in Cx. pipiens carcasses. While there is no evidence of entomopathogenicity or entomotoxicity of A. tenuissima toward mosquito larvae, one study has reported the entomotoxicity of A. tenuissima extracts toward larvae of Galleria mellonella and adult Acheta domesticus (Salimova et al., 2021). Interestingly, a fungus within the genus, Alternaria porri was enriched in Ae. albopictus carcasses (Tawidian et al., 2021), and causes mortality in green apple aphids and lowers hatch rates of European corn borer eggs (Lynch and Lewis, 1978; Sarhan and Baji, 2009). A. tenuissima was also detected in the breeding water and guts of Cx. pipiens larvae, suggesting that this fungus is ingested and may disseminate from the gut into the larval body cavity. Future studies are necessary to determine the entomopathogenic and/or entomotoxigenic potential of A. tenuissima in mosquito larvae.
In conclusion, the present study provides the first comprehensive description of the fungal community composition and assembly in Cx. restuans and Cx. pipiens mosquito larvae. Our results show that the fungal community assembly remains generally similar between the two Culex species, and suggests that the observed variation is likely due to differences in larval feeding behavior. We show that the larval breeding site is the key driver of fungal community assembly in mosquito larvae. We further show that mosquito feeding behavior and fungal mode of nutrition contribute to differences in fungal community assembly across mosquito tissues. However, the observed tissue preference was more evident in Cx. pipiens than in Cx. restuans larvae and broadly recapitulated our previous report focusing on Ae. albopictus. Future studies require greater sampling of breeding sites co-inhabited by Cx. restuans and Cx. pipiens larvae to better assess the impact of mosquito feeding behavior differences on fungal community assembly for these two species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA798403.
Author Contributions
PT performed experiments and analyzed data. KM supervised the project. PT and KM drafted the manuscript and all authors contributed to the final version of the manuscript. All authors conceived the original idea, designed experiments, and approved the submitted version.
Funding
This study was supported by funding from National Institutes of Health grant number R01AI140760, USDA-ARS specific cooperative agreement 58-5430-4-022, and USDA National Institute of Food and Agriculture Hatch project 1021223. This was contribution no. 22-132-J from the Kansas Agricultural Experiment Station.
Author Disclaimer
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
KM and PT thank all members of the Michel Laboratory for their help with insect rearing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.911085/full#supplementary-material
Footnotes
References
Abarenkov, K., Nilsson, R. H., Larsson, K.-H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Andreadis, T. G. (2012). The contribution of Culex pipiens complex mosquitoes to transmission and persistence of West Nile Virus in North America. J. Am. Mosq. Control Assoc. 28, 137–151. doi: 10.2987/8756-971X-28.4s.137
Andreadis, T. G., Anderson, J. F., Vossbrinck, C. R., and Main, A. J. (2004). Epidemiology of West Nile Virus in Connecticut: a five-year analysis of mosquito data 1999–2003. Vector Borne Zoonotic Dis. 4, 360–378. doi: 10.1089/vbz.2004.4.360
Asahina, S. (1964). Food material and feeding procedures for mosquito larvae. Bull. World Health Organ. 31, 465–466.
Badran, R. A. M., and Aly, M. Z. Y. (1995). Studies on the mycotic inhabitants of Culex pipiens collected from freshwater ponds in Egypt. Mycopathologia 132, 105–110. doi: 10.1007/BF01103782
Bascuñán, P., Niño-Garcia, J. P., Galeano-Castañeda, Y., Serre, D., and Correa, M. M. (2018). Factors shaping the gut bacterial community assembly in two main Colombian malaria vectors. Microbiome 6:148. doi: 10.1186/s40168-018-0528-y
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taberlet, P., and Kauserud, H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10:189. doi: 10.1186/1471-2180-10-189
Blaalid, R., Kumar, S., Nilsson, R. H., Abarenkov, K., Kirk, P. M., and Kauserud, H. (2013). ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 13, 218–224. doi: 10.1111/1755-0998.12065
Bokulich, N. A., and Mills, D. A. (2013). Improved selection of Internal Transcribed Spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79, 2519–2526. doi: 10.1128/AEM.03870-12
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Chavshin, A. R., Oshaghi, M. A., Vatandoost, H., Pourmand, M. R., Raeisi, A., Enayati, A. A., et al. (2012). Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi: a step toward finding suitable paratransgenesis candidates. Acta Trop. 121, 129–134. doi: 10.1016/j.actatropica.2011.10.015
Collins, F. H., and Paskewitz, S. M. (1996). A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol. Biol. 5, 1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x
Coon, K. L., Brown, M. R., and Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25, 5806–5826. doi: 10.1111/mec.13877
Coon, K. L., Valzania, L., McKinney, D. A., Vogel, K. J., Brown, M. R., and Strand, M. R. (2017). Bacteria-mediated hypoxia functions as a signal for mosquito development. PNAS 114:E5362–E5369.
Dennison, N. J., Jupatanakul, N., and Dimopoulos, G. (2014). The mosquito microbiota influences vector competence for human pathogens. Curr. Opin. Insect. Sci. 3, 6–13. doi: 10.1016/j.cois.2014.07.004
Desjardins, A. E. (2003). Gibberella from A (venaceae) to Z (eae). Annu. Rev. Phytopathol. 41, 177–198. doi: 10.1146/annurev.phyto.41.011703.115501
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366.
Duguma, D., Rugman-Jones, P., Kaufman, M. G., Hall, M. W., Neufeld, J. D., Stouthamer, R., et al. (2013). Bacterial communities associated with Culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One 8:e72522. doi: 10.1371/journal.pone.0072522
Gaio, A., de, O., Gusmão, D. S., Santos, A. V., Berbert-Molina, M. A., Pimenta, P. F., et al. (2011). Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: culicidae) (L.). Parasites Vectors 4:105. doi: 10.1186/1756-3305-4-105
Galeano-Castañeda, Y., Bascuñán, P., Serre, D., and Correa, M. M. (2020). Trans-stadial fate of the gut bacterial microbiota in Anopheles albimanus. Acta Trop. 201:105204. doi: 10.1016/j.actatropica.2019.105204
Galeano-Castañeda, Y., Urrea-Aguirre, P., Piedrahita, S., Bascuñán, P., and Correa, M. M. (2019). Composition and structure of the culturable gut bacterial communities in Anopheles albimanus from Colombia. PLoS One 14:e0225833. doi: 10.1371/journal.pone.0225833
Gardner, A. M., Anderson, T. K., Hamer, G. L., Johnson, D. E., Varela, K. E., Walker, E. D., et al. (2013). Terrestrial vegetation and aquatic chemistry influence larval mosquito abundance in catch basins. Chicago, USA. Parasites Vectors 6:9. doi: 10.1186/1756-3305-6-9
Geiser, E., Wiebach, V., Wierckx, N., and Blank, L. M. (2014). Prospecting the biodiversity of the fungal family Ustilaginaceae for the production of value-added chemicals. Fungal Biol. Biotechnol. 1:2. doi: 10.1186/s40694-014-0002-y
Gimonneau, G., Tchioffo, M. T., Abate, L., Boissière, A., Awono-Ambéné, P. H., Nsango, S. E., et al. (2014). Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol. 28, 715–724. doi: 10.1016/j.meegid.2014.09.029
Godwin, U. O., Ifeoma, U. H., Barnabas, E. -N., Chidozie, A. H., Winie, D U., and Ogechi, K. Q. (2021). Bacteria and fungi profile of mosquito oviposition sites in Eziobodo, Owerri West L.G.A., Imo State. Open Access Res. J. Sci. Technol. 1, 024–028. doi: 10.53022/oarjst.2021.1.1.0021
Jackson, B. T., and Paulson, S. L. (2006). Seasonal abundance of Culex restuans and Culex pipiens in Southwestern virginia through ovitrapping. J. Am. Mosq. Control Assoc. 22, 206–212. doi: 10.2987/8756-971X(2006)22[206:SAOCRA]2.0.CO;2
Jenkins, D. W. (1964). Pathogens, parasites and predators of medically important arthropods. Annotated List and Bibliography. Bull. World Health Organ. 30, 5–150.
Johnson, B. J., Robson, M. G., and Fonseca, D. M. (2015). Unexpected spatiotemporal abundance of infected Culex restuans suggest a greater role as a West Nile virus vector for this native species. Infect. Genet. Evol. 31, 40–47. doi: 10.1016/j.meegid.2015.01.007
Kim, C.-H., Lampman, R. L., and Muturi, E. J. (2015). Bacterial communities and midgut microbiota associated with mosquito populations from waste tires in East-Central Illinois. J. Med. Entomol. 52, 63–75. doi: 10.1093/jme/tju011
Krajacich, B. J., Huestis, D. L., Dao, A., Yaro, A. S., Diallo, M., Krishna, A., et al. (2018). Investigation of the seasonal microbiome of Anopheles coluzzii mosquitoes in Mali. PLoS One 13:e0194899. doi: 10.1371/journal.pone.0194899
Lepère, C., Domaizon, I., Humbert, J.-F., Jardillier, L., Hugoni, M., and Debroas, D. (2019). Diversity, spatial distribution and activity of fungi in freshwater ecosystems. PeerJ 7:e6247. doi: 10.7717/peerj.6247
Luis, P., Vallon, L., Tran, F.-H., Hugoni, M., Tran-Van, V., Mavingui, P., et al. (2019). Aedes albopictus mosquitoes host a locally structured mycobiota with evidence of reduced fungal diversity in invasive populations. Fungal Ecol. 39, 257–266. doi: 10.1016/j.funeco.2019.02.004
Lynch, R. E., and Lewis, L. C. (1978). Fungi associated with eggs and first-instar larvae of the European corn borer. J. Invertebr. Pathol. 32, 6–11.
Manohar, C. S., and Raghukumar, C. (2013). Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol. Lett. 341, 69–78. doi: 10.1111/1574-6968.12087
Merritt, R. W., Dadd, R. H., and Walker, E. D. (1992). Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annu. Rev. Entomol. 37, 349–374. doi: 10.1146/annurev.en.37.010192.002025
Muturi, E. J., Bara, J. J., Rooney, A. P., and Hansen, A. K. (2016). Midgut fungal and bacterial microbiota of Aedes triseriatus and Aedes japonicus shift in response to La Crosse virus infection. Mol. Ecol. 25, 4075–4090. doi: 10.1111/mec.13741
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Panzer, K., Yilmaz, P., Weiß, M., Reich, L., Richter, M., Wiese, J., et al. (2015). Identification of habitat-specific biomes of aquatic fungal communities using a comprehensive nearly full-length 18S rRNA dataset enriched with contextual data. PLoS One 10:e0134377. doi: 10.1371/journal.pone.0134377
Pereira, E., da, S., Sarquis, M. I., de, M., Ferreira-Keppler, R. L., Hamada, N., et al. (2009). Filamentous fungi associated with mosquito larvae (Diptera: Culicidae) in municipalities of the Brazilian Amazon. Neotrop. Entomol. 38, 352–359. doi: 10.1590/S1519-566X2009000300009
Rani, A., Sharma, A., Rajagopal, R., Adak, T., and Bhatnagar, R. K. (2009). Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol. 9:96. doi: 10.1186/1471-2180-9-96
Rochlin, I., Faraji, A., Healy, K., and Andreadis, T. G. (2019). West Nile Virus Mosquito Vectors in North America. J. Med. Entomol. 56, 1475–1490. doi: 10.1093/jme/tjz146
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Salimova, D., Dalinova, A., Dubovik, V., Senderskiy, I., Stepanycheva, E., Tomilova, O., et al. (2021). Entomotoxic activity of the extracts from the fungus, Alternaria tenuissima and its major metabolite, Tenuazonic Acid. J. Fungi 7:774. doi: 10.3390/jof7090774
Sarhan, A. T., and Baji, S. H. (2009). Biological control of the green apple aphids [Aphis pomi [DeGeer] (Homoptera: Aphididae) with two fungal entomopathogens. Iraqi J. Agric. Sci. 13, 149–151.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Scholte, E.-J., Knols, B. G. J., Samson, R. A., and Takken, W. (2004). Entomopathogenic fungi for mosquito control: a review. J. Insect Sci. 4:19.
Scolari, F., Sandionigi, A., Carlassara, M., Bruno, A., Casiraghi, M., and Bonizzoni, M. (2021). Exploring changes in the microbiota of Aedes albopictus: comparison among breeding site water, larvae, and adults. Front. Microbiol. 12:23. doi: 10.3389/fmicb.2021.624170
Shelomi, M. (2019). Bacterial and eukaryote microbiomes of mosquito habitats in dengue-endemic southern Taiwan. J. Asia Pac. Entomol. 22, 471–480. doi: 10.1016/j.aspen.2019.02.011
Souza, R. S., Virginio, F., Riback, T. I. S., Suesdek, L., Barufi, J. B., and Genta, F. A. (2019). Microorganism-based larval diets affect mosquito development, size and nutritional reserves in the Yellow Fever mosquito Aedes aegypti (Diptera: Culicidae). Front. Physiol. 10:152. doi: 10.3389/fphys.2019.00152
Steyn, A., Roets, F., and Botha, A. (2016). Yeasts associated with Culex pipiens and Culex theileri mosquito larvae and the effect of selected yeast strains on the ontogeny of Culex pipiens. Microb. Ecol. 71, 747–760. doi: 10.1007/s00248-015-0709-1
Tawidian, P., Coon, K. L., Jumpponen, A., Cohnstaedt, L. W., and Michel, K. (2021). Host-environment interplay shapes fungal diversity in mosquitoes. mSphere 6:e0064621. doi: 10.1128/mSphere.00646-21
Tawidian, P., Rhodes, V. L., and Michel, K. (2019). Mosquito-fungus interactions and antifungal immunity. Insect Biochem. Mol. Biol. 111:103182. doi: 10.1016/j.ibmb.2019.103182
Thongsripong, P., Chandler, J. A., Green, A. B., Kittayapong, P., Wilcox, B. A., Kapan, D. D., et al. (2018). Mosquito vector-associated microbiota: metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol. Evol. 8, 1352–1368. doi: 10.1002/ece3.3676
Timmermann, S. E., and Briegel, H. (1996). Effect of plant, fungal and animal diets on mosquito development. Entomol. Exp. Appl. 80, 173–176. doi: 10.1111/j.1570-7458.1996.tb00913.x
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian Classifier for Rapid Assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Liu, T., Wu, Y., Zhong, D., Zhou, G., Su, X., et al. (2018). Bacterial microbiota assemblage in Aedes albopictus mosquitoes and its impacts on larval development. Mol. Ecol. 27, 2972–2985. doi: 10.1111/mec.14732
Winters, A. E., and Yee, D. A. (2012). Variation in performance of two co-occurring mosquito species across diverse resource environments: insights from nutrient and stable isotope analyses. Ecol. Entomol. 37, 56–64. doi: 10.1111/j.1365-2311.2011.01337.x
Yee, D. A. (2008). Tires as habitats for mosquitoes: a review of studies within the eastern United States. J. Med. Entomol. 45, 581–593. doi: 10.1603/0022-2585(2008)45[581:tahfma]2.0.co;2
Yee, D. A., Kesavaraju, B., and Juliano, S. A. (2004). Larval feeding behavior of three co-occurring species of container mosquitoes. J. Vector Ecol. 29, 315–322.
Keywords: fungal community, Culex pipiens, Culex restuans, larvae, fungal diversity
Citation: Tawidian P, Jumpponen A and Michel K (2022) Patterns of Fungal Community Assembly Across Two Culex Mosquito Species. Front. Ecol. Evol. 10:911085. doi: 10.3389/fevo.2022.911085
Received: 01 April 2022; Accepted: 20 June 2022;
Published: 05 July 2022.
Edited by:
Terezia Horvathova, Institute of Soil Biology (ASCR), CzechiaReviewed by:
Matthew Christopher Ikaika Medeiros, University of Hawaii at Manoa, United StatesYongqi Shao, Zhejiang University, China
Copyright © 2022 Tawidian, Jumpponen and Michel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristin Michel, kmichel@ksu.edu
 Patil Tawidian
Patil Tawidian Ari Jumpponen
Ari Jumpponen Kristin Michel
Kristin Michel