Tropical Palm Endophytes Exhibit Low Competitive Structuring When Assessed Using Co-occurrence and Antipathogen Activity Analysis
- 1Evolution et Diversité Biologique (EDB), Université Toulouse 3 Paul Sabatier, CNRS, IRD, UMR 5174, Toulouse, France
- 2CNRS, Institut de Chimie des Substances Naturelles, UPR2301, Université Paris Saclay, Gif-sur-Yvette, France
- 3Sorbonne Université, CNRS, Laboratoire de Biodiversité et Biotechnologies Microbiennes, USR3579, Observatoire Océanologique, Banyuls-sur-Mer, France
Leaf-inhabiting fungal and bacterial endophytes are at their most diverse in tropical rainforest plant hosts, with some influencing host plant fitness as either symbionts or pathogens. Endophyte activity and community composition is thought to depend on competition amongst co-occurring species for resources. Here, we reveal the strength of competitive interactions between endophytes by combining e-DNA metabarcoding to characterize the community with culturing assays to ascertain their potential activity. The endophyte community associated with the understory palm Astrocaryum sciophilum was determined by extracting eDNA from 120 leaflets of eight plants located in a lowland rainforest in French Guiana. This revealed 516 fungal and 606 bacterial Operational Taxonomic Units (OTUs). Co-occurrence analysis of the most abundant OTUs revealed that direct fungal-fungal interactions were overall more negative than bacterial-bacterial interactions. This was confirmed by C-score calculations for the whole endophyte community, revealing significantly greater levels of species segregation for fungi but not bacteria when compared with simulated random communities. Following the culturing of 131 fungal, and 66 bacterial Astrocaryum endophytes collected from the same leaves, Minimal Inhibitory Concentrations of ethyl acetate culture extracts were evaluated against the fungus Trichophyton rubrum and the bacterium Staphylococcus aureus. Overall, a minority of extracts demonstrated antipathogen activity, with greater antifungal activity expression for both fungi and bacteria when compared with antibacterial activity. In order to explore whether this activity translated into competitive structuring of the community, a BLAST was conducted to match metabarcoding sequences with Sanger derived culture sequences. This demonstrated that cultures displaying antipathogen activity were significantly more likely to co-occur with fungi if they were bacteria, and bacteria if they were fungi. Nevertheless, overall correlation values averaged around zero. These results indicate that for Astrocaryum palms, fungal endophytes are likely to play a greater role in determining colonization success of subsequent endophytes whether these be fungal or bacterial, but that overall endophyte communities do not display strong patterns of community structuring through competition. Endophyte communities are thus likely to reach relative stasis in mature leaves, where the diverse community of fungi and bacteria amongst other factors prevent the subsequent establishment of pathogens.
Introduction
Tropical forests have been shown to support some of the highest levels of foliar endophyte diversity (Arnold and Lutzoni, 2007; Griffin and Carson, 2018), with advances in the field dramatically increasing rates of their detection (Izuno et al., 2016; Haruna et al., 2018). These organisms colonize newly flushed leaf tissues (Arnold et al., 2003; Herre et al., 2007), before establishing a symbiotic relationship with their host plant (Carroll, 1988), ranging from mutualistic (Arnold and Engelbrecht, 2007; Herre et al., 2007; Bittleston et al., 2010; Christian et al., 2019), through commensualist (Arnold and Herre, 2003), to pathogenic (Saikkonen et al., 1998; Arnold et al., 2000; Rodrigues et al., 2010; Griffin et al., 2016). Studies of endophyte activity and demonstrations of their symbiotic properties are often performed under simulated conditions in vitro, in the absence of complex interactions likely to occur in a more natural endophyte communities and mature leaves (Crowther et al., 2018). As such, they may over represent the role of certain endophytes as either pathogens or symbionts.
The ability of endophytes to colonize leaves depends on host plant identity and dispersal limitation following environmental gradients (Cordier et al., 2012; Zimmerman and Vitousek, 2012; Higgins et al., 2014; Solis et al., 2016; Vincent et al., 2016). Yet the role of competition between co-occurring endophytes within each leaf is likely to be as important in determining colonization success, with a “priority effect” observed in microbial taxa colonizing phyllospheres (Maignien et al., 2014; Vacher et al., 2016). Competition for limited resources within a community of organisms is thought to determine the realized niche of each species (Chase and Leibold, 2003). Specifically, for endophytes, the species inhabiting a leaf are likely to limit the subsequent colonization and activity of others (reviewed in Van Bael et al., 2017). Endophytes have been shown to confer a defense against pathogens (Arnold et al., 2003; Herre et al., 2007), and although these findings are based on endophyte communities of low complexity, it is likely that a plant's established microbiome will compete with a colonizing pathogen (Gazis and Chaverri, 2015). Similarly, Kennedy et al. (2009, 2010) demonstrate priority effects by initial colonists preventing further colonization of roots by fungi in ectomycorrhizal systems, indicating that competition may have important implications in analogous endophyte communities.
Assessing interactions between species in ecological communities can be achieved by constructing bipartite networks, which are then quantified using co-occurrence analysis (Dormann et al., 2008). Microbial association networks typically contain positive links which indicate species co-occurrence, and negative links which indicate co-exclusion. These associations can arise from direct interactions between species (Jakuschkin et al., 2016), although are also likely to occur as a result of indirect pathways in more complex communities. At the community level, checkerboard-score (C-score) calculations provide a viable alternative to simple co-occurrence, since they compare observed patterns in community assemblages with randomized null communities (Gotelli, 2000). In doing so, C-score calculations reveal potential trends in species competitive exclusion or conversely aggregation, which can be further studied using more thorough methods of network analysis (Barberán et al., 2012; Williams et al., 2014).
Two primary methods are employed to characterize endophyte communities (Sun and Guo, 2012). Initial endophyte research largely focused on culturing of isolated strains of fungi and bacteria on agar plates (see reviews by Griffin and Carson, 2015, 2018). Although this approach is limited to endophytes capable of growing in culture (Hyde and Soytong, 2008), it has enabled calculations of potential activity, through co-culturing of competing endophytes, or in the presence of antibiotics (e.g., Casella et al., 2013). In addition, this approach has revealed the potential mechanisms by which competition may occur, through the production and isolation of bioactive compounds (Bertrand et al., 2013, 2014). For example, André et al. (2017) examined the role of the fungal endophyte Colletotrichum gloeosporioides which colonizes tropical fruit-bearing plants such as mango and papaya, and which under certain circumstances develops as a pathogen. In isolating biotic compounds produced by this endophyte, they were able to demonstrate potential antipathogenic mechanisms. More recently, next generation sequencing (NGS) of nucleic acids from environmental samples has been developed to determine species occurrence (Zinger et al., 2016; Taberlet et al., 2018). These methods rely on the extraction of environmental DNA (eDNA), using amplification primers which are specific to the taxa of interest, and as such will capture any trace of bacterial or fungal DNA within host plant leaves before amplifying these to enable their sequencing and subsequent identification using sequence databases. This approach has enabled a wealth of studies which reveal the cryptic diversity of endophytes and pathogens, with the high number of co-occurring species detected suggesting complex networks of potential interactions. However, although these methods are able to determine association, they struggle to demonstrate explicit ecological interactions (Jakuschkin et al., 2016; Bohan et al., 2017). The inferred effects of community structure can only go so far in evoking functional processes, which depend on tests of potential activity in culture (Crowther et al., 2018).
In order to determine whether competition between co-occurring endophytes observed in natural communities matched with their potential activity in culture, we combined these two approaches, focusing on the role of established endophytes competing with fungal and bacterial pathogen colonizers. We selected the tropical understory palm Astrocaryum sciophilum (Miq.) Pulle as a model host to test for patterns in endophyte colonization. By trapping significant quantities of leaf litter within their leaves over long lifespans, these plants not only concentrate nutrient release (Alvarez-Sanchez and Guevara, 1999; Alvarez-Sanchez et al., 2016), but are also likely to increase their exposure to endophyte spores. In harboring a more diverse endophyte community, this species demonstrates great potential for revealing patterns of competition and co-occurrence which may be more difficult to detect in shorter lived species. Furthermore, the palm is characteristic of undisturbed primary forest, and its growth strategy of producing a new leaf every 18 months makes sampling of age classes and especially old leaves possible (Charles-Dominique et al., 2001, 2003).
Our study aims to describe the bacterial and fungal endophyte communities of Astrocaryum sciophilum and investigate their interactions, in order to reveal their potential role in competitive exclusion of pathogens. This was achieved by exploring co-occurrence patterns derived from NGS, and antipathogenic activity determined through Minimal Inhibitory Concentration evaluations. We hypothesized that (i) competition would be higher in endophytic communities than predicted by null models, (ii) bacteria and fungi would show more negative interactions between each other, and (iii) that strains showing high antipathogenic activity would co-occur less with other microorganisms than strains with low antipathogen activity.
Materials and Methods
Sample Location
Sample collection took place on the Saint Elie track (5°17′35.0″N, 52°02′55.1″E), ~10 km from the coastal town of Sinnamary in French Guiana. Over 1 ha of pristine forest, eight Astrocaryum sciophilum were detected and sampled. Depending on their height, two to eight leaves were accessible per individual (mean = 5), and three leaflets were collected per leaf. All collected leaves were a minimum of 1 year old ranging up to 14 years, chosen so as to avoid visible signs of pathogen or herbivore damage. In total 120 leaflets we collected. Initial sample preparation for e-DNA extraction and culture collection took place in Institut Pasteur in Cayenne in 2014. The same trees were further resampled in 2015 in order to increase the number of cultured endophytes.
Sample Design and Preparation
In order for subsequent analysis to focus only on the endophyte community, upon collection, leaflets were immediately transported to a field laboratory where they were surface sterilized following a modification of the protocol detailed by Weber et al. (2004). Firstly, leaflet surfaces were cleaned and scraped using scourers, before immersion in 70% aqueous ethanol (3 min) followed by 5% aqueous bleach (5 min) and finally in water (1 min). One fragment of 1 cm2 was cut with a sterile razor blade within each leaflet. Each fragment was cut in half. One half to be used to sequence the endophytic community was dried at 45°C for 3 days, whilst the second half was kept fresh and placed on petri dishes to allow endophyte growth. During a second sampling campaign, leaflet fragments were only processed for culture on petri dishes.
DNA Isolation, PCR Amplification and Sequencing
Dried leaflet fragments were placed into a 96 well plate, and a 3 mm aluminum bead was added to each well. To facilitate the grinding step, the sealed plates were placed in a −80°C freezer for at least an hour before grinding using a Tissue Lyser (Qiagen, Germany) for 45 s at 30 Hz, followed by centrifugation for 30 s at 6,000 × g. DNA was extracted from samples using a NucleoSpin Plant II (96) kit (Macherey-Nagel, Düren, Germany) according to the manufacturer protocol modifying only cell lysis time to 1 h and elution volume to 100 μL.
PCRs were performed using two primers; ITS1 nuclear rDNA primers to target fungi [Fwd: ITS5 GGAAGTAAAAGTCGTAACAAGG (Epp et al., 2012) and a modified version of Rev: 5.8S_Fungi CAAGAGATCCGTTGTTGAAAGTK, Taberlet et al., 2018], and 16S rDNA (V5-V6) primers to target bacteria (Fwd: GGATTAGATACCCTGGTAGT and Rev: CACGACACGAGCTGACG, Fliegerova et al., 2014). To discriminate samples after sequencing, forward and reverse primers were synthetized with a combination of two-different 8-nucleotide tags per sample. As such, each DNA extract was amplified with a unique combination of tagged primers. Each PCR reaction was performed in a total volume of 20 μl and comprised 10 μl of AmpliTaq GoldMaster Mix (Life Technologies, Carlsbad, CA, USA), 5.84 μL of nuclease-free water Ambion™ (Invitrogen, Waltham, Massachusetts, USA), 0.25 μM of each primer, 3.2 μg of BSA (Roche Diagnostic, Basel, Switzerland), and 2 μl of DNA template. Two PCRs per sample were performed under the following conditions: Polymerase reactivation 10 min at 95°C, followed by 35 (fungi) or 30 (bacteria) cycles of 30 s at 95°C, 30 s at 55°C (fungi), or 57°C (bacteria) and 1 min at 72°C; followed by a final step of 7 min at 72°C. Two wells in each plate were filled with water, to act as PCR negative controls. Amplicons were pooled and libraries were prepared with the TruSeqNano PCR free Illumina kit and were sequenced on Paired-end (2X250 bp) in the Illumina Miseq platform (Illumina, San Diego, CA, USA) at the INRA Genotoul-GetPlaGe core facility (Toulouse, France) using the Paired-end MiSeq Reagent Kit V3 (Illumina, San Diego, CA, USA), following the manufacturer's instructions.
Sequence Data Curation
The generated reads were subject to a data-curation pipeline using scripts in R (R Core Team, 2013), and the OBITools package (Boyer et al., 2016). Firstly, paired-end read assembly was conducted, before reads were assigned to samples, and were dereplicated. Low-quality sequences (with a pair alignment score of 50 or below) and singletons were removed prior to further analysis. Pairwise distances between the remaining sequences were then calculated using the sumaclust function, which determines pairwise sequence dissimilarity using the raw number of mismatches. Sequence clusters with similarity of 97% or above were defined as operational taxonomic units (OTUs) which are commonly used as proxies for species during molecular analysis, with this 97% threshold used in the majority of fungal studies (Coissac et al., 2012).
Taxonomic assignment of OTUs was conducted using reference databases with the ecotag function, which uses global alignment of sequences against full-length references. For fungi the reference database was obtained by running an in-silico PCR with the ecoPCR program (Ficetola et al., 2010) on Genbank (release 197; https://www.ncbi.nlm.nih.gov/genbank/) using the primer pairs employed here. Taxonomic assignment yielding the highest similarity score was kept, with similarity scores for Fungi ranging from 0.5 to 1 with sequences in the National Center for Biotechnology Information (NCBI) database. For Bacteria, taxonomic assignment was conducted using the SILVA database (Quast et al., 2012), with similarity parameters kept at 97%.
OTUs which could represent contaminants arising during extraction (present within the extraction negative controls) and during PCR (present within PCR negative controls) were removed from the dataset by comparing occurrence in samples. The influence of chimeras formed during PCR and sequencing was reduced by comparing occurrence in samples with that in the blank controls, detecting for non-used tag combinations, as follows. Potential contaminant OTUs were detected by calculating the mean sum of reads in blanks, divided by the mean sum of reads across both blanks and samples. For each OTU, the acceptable level of OTU abundance present in blanks was set to be 5% or less than the total number of reads. Above this threshold, an OTU was more likely to be a contaminant, and as such it was excluded. OTUs occurring only in negative controls were also removed, with their random distribution across plates representative of any cross-contamination of genetic information. This resulted in the removal of 137 bacterial (833 reads) and 140 fungal OTUs (2,457 reads).
All OTUs which were identified as non-fungal following taxonomic assignment were removed from the fungal database. Bacterial sequences which were identified as originating from ribosomes or chloroplasts were removed from the bacterial dataset, since these likely represented plant host genetic material rather than that of bacterial endophytes (representing 2,140,196 reads). We excluded fungal samples with <100 reads from subsequent analysis. These low-quality samples likely result from PCR amplification failure. This filter was chosen as a result of the majority of samples averaging a much higher read count. This resulted in the removal of 27 samples, with subsequent analysis for the fungal communities performed on a dataset of 73 leaflets. In comparison, bacterial read counts per sample were much lower, due to the dominance of ribosome and chloroplast data. Once these were removed, all samples containing a minimum of 1 read were kept in the dataset, resulting in the removal of one leaflet, with subsequent analysis for the bacterial communities performed on a dataset of 119 leaves.
Following clustering of sequence reads with a 97% similarity cut-off, and removal of contaminants, 554 OTUs (400,548 reads) were obtained for fungi, and 606 OTUs for bacteria (14,034 reads). Subsequent analysis was performed on these reduced datasets.
Isolation in Culture and Identification of Endophytes
Culturing methods followed those detailed by Casella et al. (2013), briefly outlined below. After surface sterilization, leaves were cut into small pieces (1–0.5 cm2) which were placed on Potato Dextrose Agar medium (PDA, Fluka Analytical, Germany) in Petri dishes at 28°C (4–5 parts per Petri dishes). Each individual hyphal tip of emerging fungi was removed and placed on a sterile PDA culture medium in a 10 cm Petri dish. The leaf fragments were cultured for a maximum of 1 month. All the isolated endophytic strains were deposited in the ICSN/CNRS Strain library (France). The strains are maintained in triplicate in 2 ml Eppendorf tubes containing 1 ml of a solution of glycerol and water (1:1) at −80°C.
DNA from cultures was extracted using Whatman® FTA Card Technology. Fungal and bacterial strains were identified using Polymerase Chain Reaction (PCR). Internal Transcriber Spacer (ITS) regions were amplified using universal primers ITS1F and ITS4 (White et al., 1990) and sequencing were carried out with ITS4. To amplify 16S regions, universal primers 27Fmod and 1492Rmod were used. Sequencing of 16S regions were carried out with the inter 907R primer. PCR was conducted at the Observatoire Oceéanologique de Banyuls as detailed in Fagervold et al. (2013). Obtained sequences were deposited on the NCBI database (see Tables S1–S4 for taxonomic identification and accession numbers).
Culture and Extraction
Each strain was cultivated at 28°C in 10 Petri dishes (10 cm diameter) containing PDA culture media. The culture was then extracted with ethyl acetate (EtOAc) at room temperature for 24 h. The organic phase was removed via filtration, washed three times with H2O, dried with anhydrous solid Na2SO4 and evaporated using a rotary evaporator under reduced pressure to yield a crude mixture.
Antimicrobial Activities
The ATCC strain was purchased from the Pasteur Institute and the clinical isolate was provided by Phillipe Loiseau (University Paris Sud, Châtenay-Malabry, France). The strains used in this study were methicillin-resistant Staphylococcus aureus (MRSA) ATCC33591 and a clinical isolate of Trichophyton rubrum (strain SNB-TR1). Extracts were tested according to the reference protocol of the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2016). The standard microdilution test as described by the Clinical and Laboratory Standards Institute guidelines (M7-A8 and M38-A2) was used to determine minimal inhibition concentrations (MIC; Rodrigues et al., 2010; Nirma et al., 2013). Briefly, crude extracts were tested at concentrations ranging from 256 to 8 μg/mL. The microplates were incubated at 35°C, and MIC values were calculated after 24 h for MRSA and 5 days for T. rubrum. The MIC values refer to the lowest concentration preventing visible growth of the pathogen in the wells. All assays were conducted in duplicate. Vancomycin and fluconazole (2–1 μg/ml) were used as positive controls for the assays. Antimicrobial standards were purchased from Molekula (Dorset, UK).
Following Rodrigues et al. (2019), to allow for direct regression of analytical data as a function of antipathogen potential, a score for both antifungal and antibacterial activity were attributed to each extract. A MIC >256 μg/mL was noted as 0, of 256 μg/mL as 1, of 128 μg/mL as 2, of 64 μg/mL as 3, of 32 μg/mL as 4, of 16 μg/mL as 5, and of <8 μg/mL as 6.
In order to combine identifications of sequenced OTUs with cultured species, we performed a local BLAST of the NGS derived sequence database with the culture derived sequence database (Altschul et al., 1997). Here, sequences were only considered matches if they had above 97% similarity to follow OTU cluster dimensions, and if they had 80 or 200 base pairs or more in common for bacteria and fungi, respectively.
Statistical Analysis
All statistical analysis was performed using R version 3.6.1 programming language (R Core Team, 2013).
To quantify endophyte species richness and diversity, rarefaction curves for fungi and bacteria were compiled using the “iNEXT” package (Hsieh et al., 2016). These curves demonstrated whether sampling was sufficient in representing the diversity of Astrocaryum sciophilum endophyte diversity, defined by whether curves reached asymptote. To determine taxonomic diversity of sampled endophytes, each OTU was classed at order level, and their relative occurrence was calculated. This revealed which orders occurred more frequently.
In order to detect patterns in OTU co-occurrence, potential competition and exclusion, three approaches were applied. Firstly, reduced datasets for both bacteria and fungi were prepared, consisting of only the 100 most abundant OTUs (across 119 leaves for bacteria, and 74 leaves for fungi). This reduced dataset allowed for statistical testing of OTU correlations, which would not be possible for the rarest OTUs that would otherwise overestimate the rates of exclusion and subsequent C-scores. For each of these, coefficients of correlation were determined, using the bipartite and corrplot packages (Dormann et al., 2008; Wei et al., 2017). This revealed which OTUs were positively or negatively correlated, but also the overall tendency of the community to demonstrate competitive or neutral organization. In order to see whether patterns changed when considering fungi and bacteria together as one community, the analysis was repeated on a dataset combining the most common fungi and bacteria, using only data from the 74 leaves available for both. This dataset consisted of 91 fungal and 9 bacterial OTUs.
Next, in order to confirm the presence of competition, a community level C-score was calculated for both bacteria and fungi, using the EcoSimR package (Gotelli and Ellison, 2013). Here, the community composition was compared with 1000 random permutations of the dataset to create a null distribution curve. An observed c-score which is significantly higher than a null distribution is considered to be structured by segregation, significantly lower structured by aggregation, and of no significant difference to experience no competition.
Finally, in order to determine whether cultured strains displaying antipathogen activity influenced patterns of community assembly, we compared the determined correlation coefficient for each cultured bacteria and fungi with the whole community. The difference between fungal and bacterial responses was calculated with an ANOVA using the “aov” function in R, with normality confirmed using the shapiro-wilk test.
Results
Bacterial and Fungal Composition
Based on sequencing, leaflets were found to contain an average of 18 bacterial (range: 4–191) and 22 fungal OTUs (range: 4–161). In total, these amount to 554 OTUs (400,548 reads) for fungi, and 606 OTUs for bacteria (14,034 reads). Saturation curves suggested that our sampling was comprehensive for fungi, but that bacterial diversity was far from being representative (Figure 1). Using SILVA and NCBI to assign taxonomy to our OTUs resulted in over 99% of bacterial OTUs being assigned to order level, in comparison to only 38% of fungal OTUs (Figure 2). The three most common orders were Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria in the case of bacteria, and Agricales, Capnodiales, and Hypocreales for fungi.
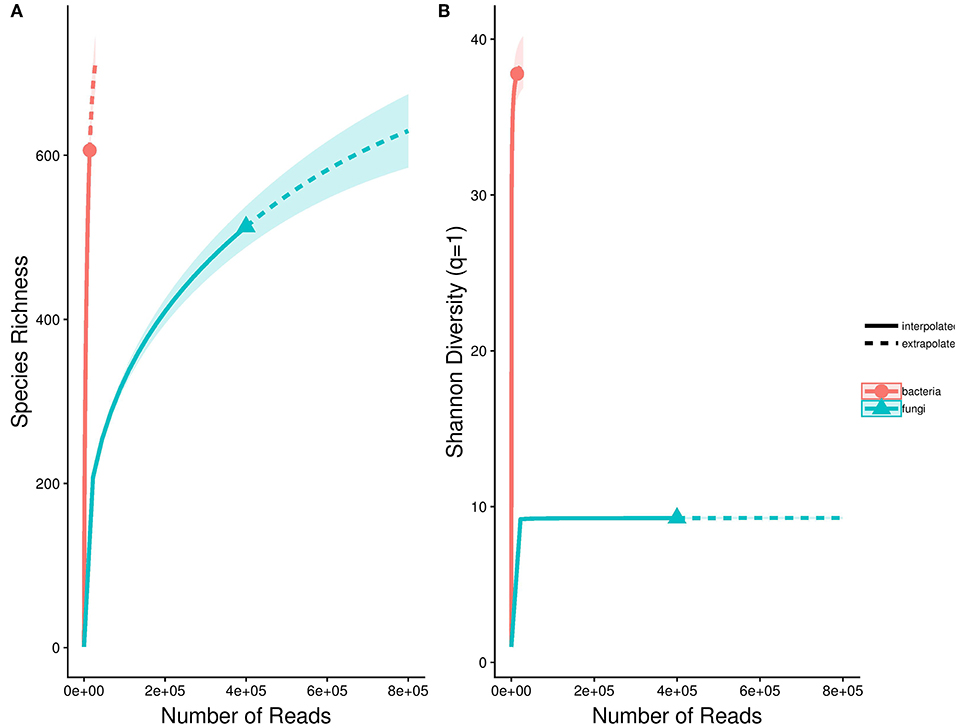
Figure 1. Species accumulation curves for bacterial and fungal endophytes detected in the sample pool. These are represented using abundance-based Hill numbers, which take rare sequences into account through a scaling parameter of diversity termed q. (A) Richness (q = 0), (B) Shannon diversity (q ~ 1). Dashed line indicates projected richness.
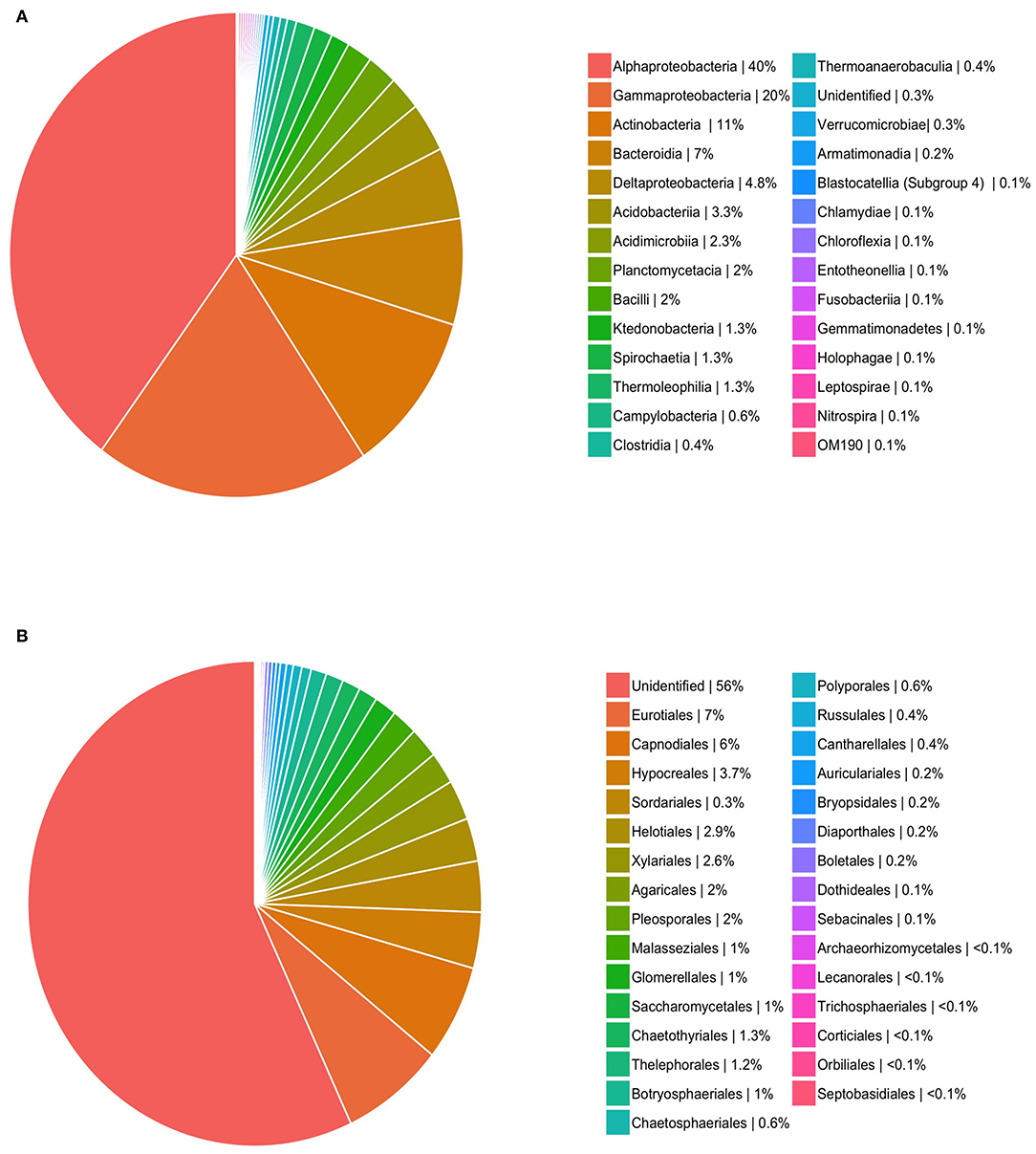
Figure 2. Relative occurrence (i.e., number of times each order occurs in sampled leaves out of total pool of occurrences) for OTU orders for (A) bacteria and (B) fungi.
Bacterial and Fungal Co-occurrence
A coefficient of correlation for co-occurrence was calculated for each isolated bacterial and fungal OTU. Average bacterial co-occurrence correlation was 0.273 ± 0.23 (mean ± SD), whilst average fungal co-occurrence correlation was 0.062 ± 0.19. When datasets were combined, leaf endophyte communities consisting of both bacteria and fungi had an average co-occurrence correlation of 0.233 ± 0.13. ANOVA demonstrated that bacterial correlation is significantly more positive in comparison to fungi [F(1, 19998) = 5,026, Pr(>F) = <2e-16], with a trend toward positive correlation for bacteria in comparison to more neutral association for fungi (Figure 3). Calculation of C-scores demonstrated that bacterial communities are not significantly different in their arrangement than those recorded as occurring at random, suggesting no apparent segregation or aggregation as a result of competition. Fungal community composition, on the other hand, demonstrated significantly more segregation than simulated null communities, as did the dataset which contained both fungi and bacteria combined (Figure 4).
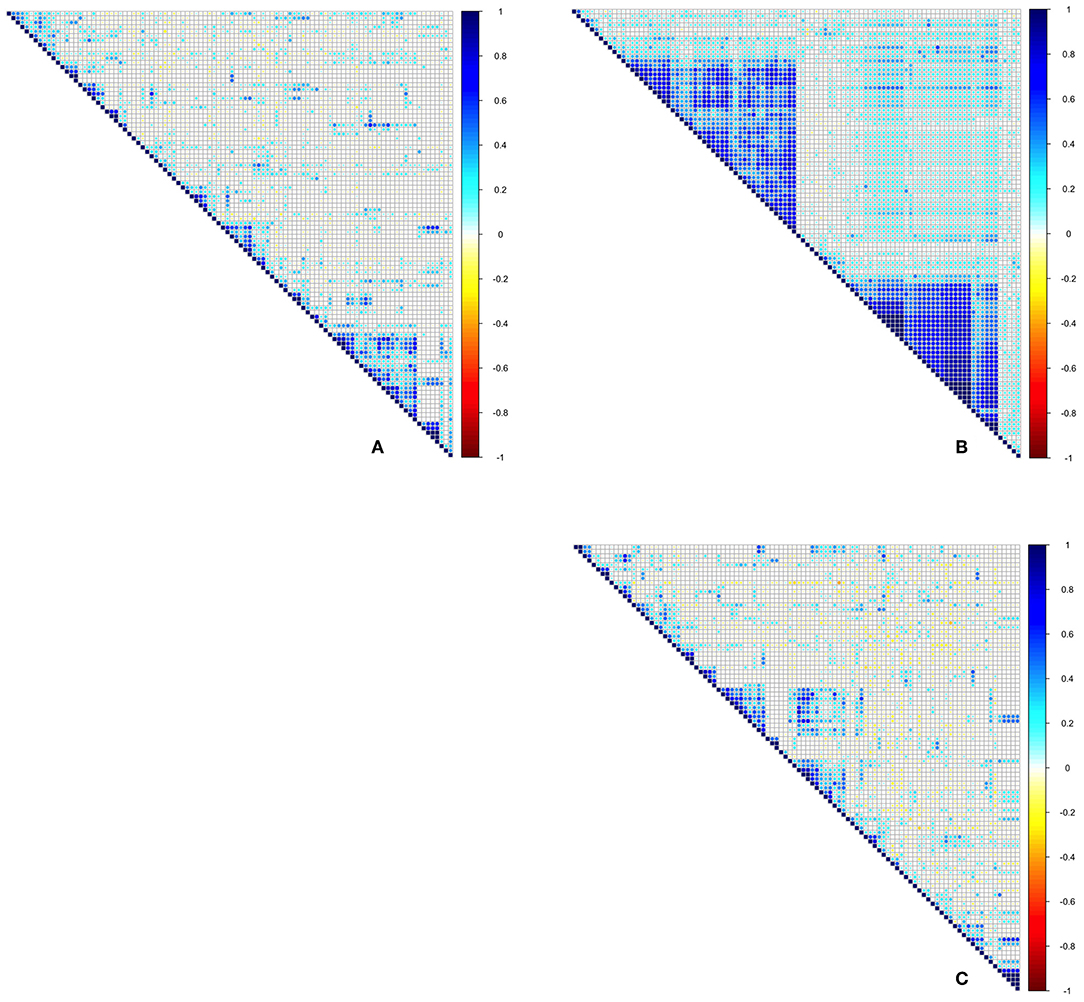
Figure 3. Spearman correlation plots for the 100 most abundant OTUs for (A) Bacteria-Bacteria, (B) Fungi-Fungi, (C) Bacteria-Fungi. Colouration from blue to red indicates whether OTUs consistently co-occur across samples through to whether OTUs consistently never occur together.
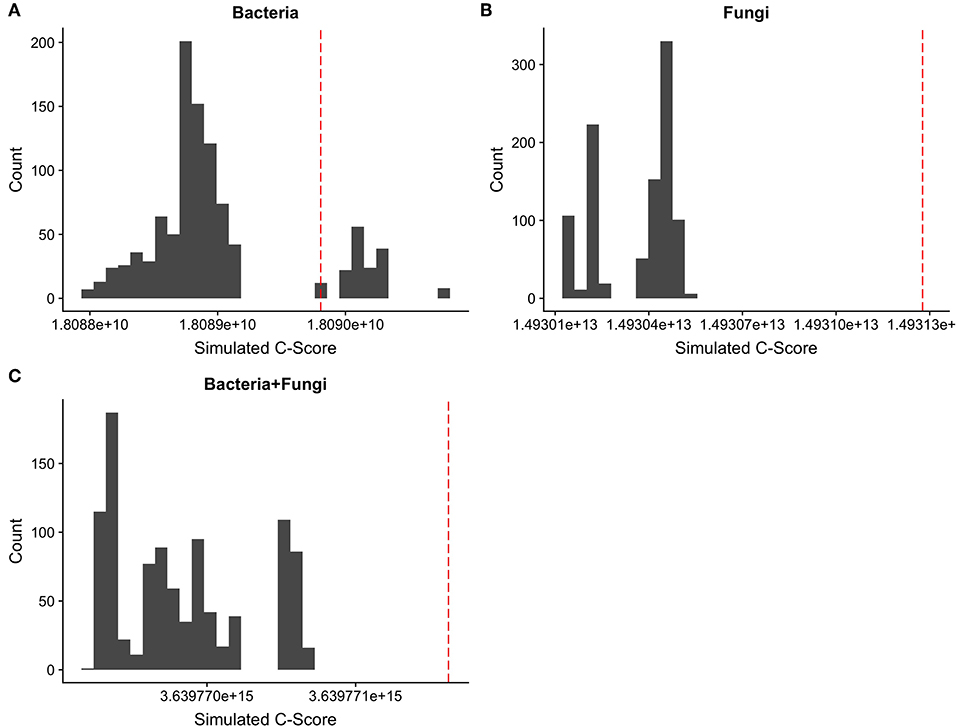
Figure 4. Distribution of c-scores for 1,000 random permutations of (A) bacterial, (B) fungal, and (C) combined OTU datasets. The red line indicates the observed c-score for each OTU community. An overlap of the observed c-score with permutated c-score indicates no difference of c-score from that generated at random, whilst an observed c-score much higher than the permutated data indicates significant segregation effects between OTUs and thus potential competition.
Cultures and Their Activity Levels
A total of 66 bacterial, and 131 fungal cultures were generated from samples, representative of a range of orders, although dominated by Bacillales, Burkholderiales, and Enterobacteriales for bacteria and Botryosphaeriales, Glomerellales and Xylariales for Fungi (Tables S1, S2). Only a minority of these cultures displayed antipathogenic activity; of the 66 bacteria, 7 displayed antibacterial, and 23 displayed antifungal properties. Of the 131 fungi, 16 displayed antibacterial and 67 displayed antifungal activity. Culture Activity Scores revealed that the strength of antipathogen effects varied, with only a limited number of cultures displaying strong competitive effects (Figure 5; Tables S3, S4).
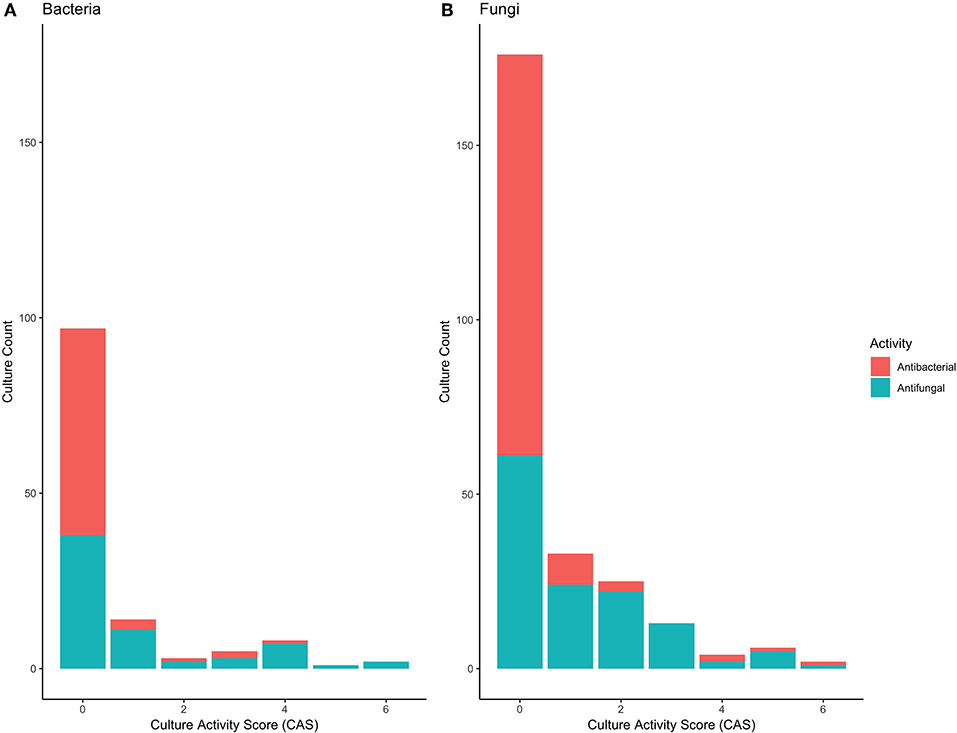
Figure 5. Antipathogen activity scores for (A) 66 bacterial, and (B) 131 fungal isolates, based on the measured minimum inhibitory concentrations (MIC sensu, Rodrigues et al., 2019). Both antifungal and antibacterial activity for each isolate was determined. A value of 0 indicates no inhibition and thus no antipathogen activity, whilst a value of 6 indicates high inhibition and thus antipathogen activity.
Combining NGS and Culture Data to Determine Correlation Effects
BLAST analysis revealed that aligning isolated cultures with OTUs defined by NGS was relatively ineffective, with only 59 of the 197 cultures matching, corresponding to 13 OTU sequences (8 for bacteria, 5 for fungi). ANOVA of coefficients of correlation revealed that bacteria which displayed antipathogen activity (both antibacterial and antifungal) in culture tended to correlate more positively with the fungal community than with bacterial community (Table S5). Similarly, fungi which displayed antipathogenic activity in culture tended to correlate more positively with bacterial than with fungal community (Figure 6). Nevertheless, mean correlation of community co-occurrence for each of the cultures remained centered around zero.
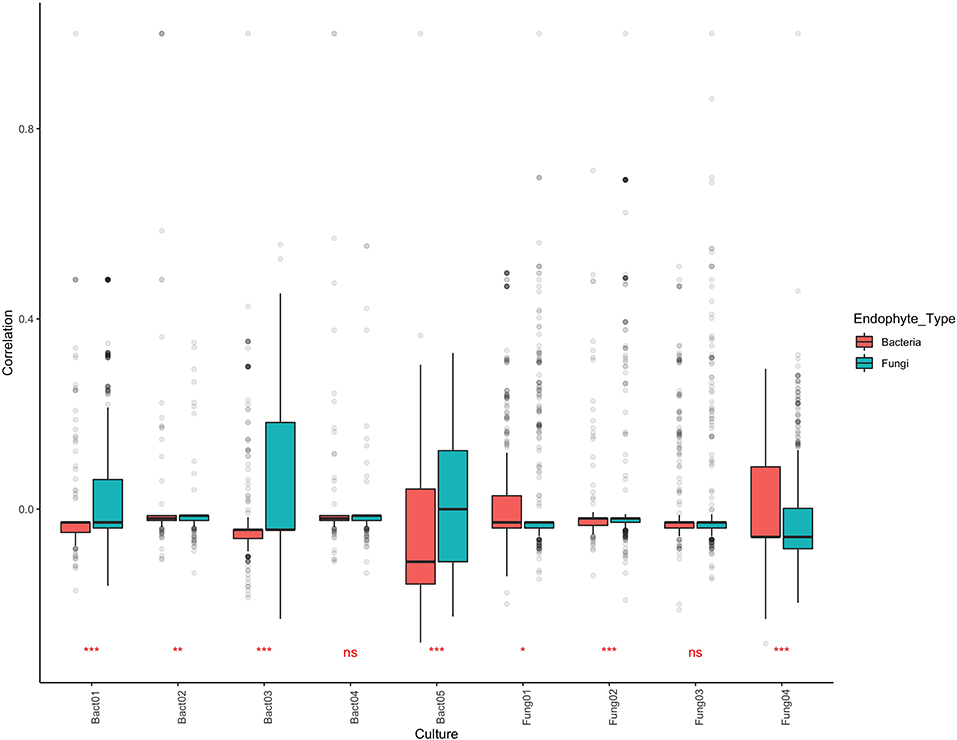
Figure 6. Community co-occurrence correlation values for nine endophytes which demonstrated antipathogen activity when cultured corresponding to OTUs in the metabarcoding dataset. Values of 1 indicate consistent co-occurrence and −1 indicate OTUs never co-occurring. Significance values shown indicate whether community co-occurrence correlation values for the bacteria differ from those of fungi.
Discussion
In spite of high levels of co-occurrence, abundance and diversity, endophyte communities in Astrocaryum leaves do not appear to be structured by competition, exhibiting minimal potential activity against pathogenic fungi and bacteria. Using NGS techniques to capture greater levels of the cryptic endophyte diversity than that revealed through culturing methods and determine co-occurrence confirmed a lack of strong competitive interactions. Using culturing techniques to isolate endophytes, and test their competitive effect against laboratory strains of fungi and bacteria confirmed varying levels of antipathogen activity. Contrary to our hypothesis, the presence of OTUs demonstrating high levels of antipathogen activity in culture did not result in modified community structure as a result of competitive effects. Overall, the study demonstrates high levels of co-occurrence within endophyte communities, with relatively low levels of competition within palm leaves. Nevertheless, it has also revealed that under isolated conditions, certain endophytes display relatively high antipathogen activity.
As with other plants sampled in tropical forests, A. sciophilum leaflets contain a community of endophytes characterized by high diversity from a range of taxonomic orders. Through NGS techniques, leaf sampling has enabled representative characterization of the fungal community, although this is not the case for bacteria. For these, a large proportion of the sequences amplified using the 16S primer corresponded to genetic material of mitochondrial and chloroplast origin, potentially masking the true diversity of bacteria during amplification. This is perhaps unsurprising when extracting DNA from a plant host, further justifying a joint analysis of endophyte communities using both NGS and culturing techniques, but also the use of primers tailored to amplification of bacteria within plant tissues (e.g., Beckers et al., 2016). Culturing of endophytes was also more successful for fungi than bacteria, suggesting that this relative difference in their abundance in plant leaves is not an artifact of extraction and amplification bias. In spite of their lower number, taxonomic identification was more comprehensive for bacteria than fungi, in all likelihood due to the relatively low number of neotropical fungal sequences in global databases such as NCBI. There is a pressing need to produce more reference sequences for fungi in the tropics (Truong et al., 2017), with endophytes in particular underrepresented in fungal taxonomy. Nevertheless, these results show that over a third of identified bacterial OTUs are Alphaproteobacteria, whilst for fungi there is less dominance of specific orders. Interestingly, these leaves appear to harbor a community of endophytes which differ in the relative abundance of OTUs of other trees sampled in similar habitats (Rodrigues, 1994; Fróhlich et al., 2000; Cannon and Simmons, 2002). In particular they are not dominated by Xylariales which are often cited as the most abundant family of foliar endophyte (e.g., Arnold and Lutzoni, 2007). Overall, the identification of bacterial and fungal endophytes increases our understanding of the high levels of diversity within tropical systems, particularly important for bacteria where relatively few studies have been conducted (Griffin and Carson, 2015).
Through co-occurrence analysis of the 100 most abundant OTUs for both bacteria and fungi, it was found that overall exclusion was low with no observed strong competitive exclusion. Bacterial communities in particular demonstrate greater levels of co-occurrence. The strength of this relationship was confirmed by comparing the co-occurrence structure of total observed endophyte community with random permutations of the data to represent a null model. The calculated C-score confirms that whilst bacterial communities do not appear to be more segregated or aggregated than communities composed at random, fungal communities do demonstrate a significant level of segregation, suggesting more pronounced if low intensity competition within this community. In our analysis, we attempted to characterize the interaction occurring between fungi and bacteria, since within leaves these two communities are likely to depend on the same resource pool. When combining datasets to include the 100 most common fungal and bacterial OTUs, simulated C-score is lower, although given this community is largely composed of fungi (91 vs. 9 bacteria), this mostly represents fungal-fungal co-occurrence rather than bacterial-fungal. The absence of strong competitive effects is initially surprising, given the high numbers of different species occurring within a defined habitat of limited resources. Nevertheless, such a finding is not unique, matching those of Jakuschkin et al. (2016), who demonstrated that communities of endophytes in temperate oak tree leaves exhibited largely mutualistic, facilitative, and commensalism interactions. The diverse range of endophyte types detected are likely to represent a diverse range of life strategies, with stabilizing mechanisms enabling coexistence and resource partitioning (Chesson, 2000). A further explanation for the lack of observed competition here is that competition is likely to be at its most fierce in young leaves during initial colonization and community establishment (e.g., Arnold and Herre, 2003). Since these samples represent leaves which are at least 18 months old, much of the competition which structures the community has potentially already occurred. Future studies of competitive dynamics in endophyte communities should take this temporal aspect into account, perhaps through the repeat sampling of the same individual, and leaflets from, the same leaf through time, and observing potential shifts in c-scores.
In comparison to the total 606 bacterial OTUs and 554 fungal OTUs detected using metabarcoding of leaf e-DNA, a smaller but nevertheless substantial number of cultures (66 bacterial, 131 fungal) were established. The majority of these demonstrated no antipathogen activity. Such an absence of competitive effects mirrors the overall trend for co-occurrence within our NGS dataset. A potential explanation could be that the majority of endophytes are in fact adapted to co-occur in leaves, or that there is sufficient resource partitioning inspire of the high levels of co-occurring species. For those cultures where antipathogen activity was detected, this generally matched the observed trend for greater co-occurrence in bacteria than in fungi. Specifically, for both fungal and bacterial cultures, there was little expression of antibacterial activity, with both displaying greater levels of antifungal activity. In addition, a greater proportion of fungal cultures displayed activity when compared to bacteria.
BLAST analysis revealed that aligning isolated cultures with OTUs defined by NGS was relatively ineffective, with only 59 of the 197 cultures matching, corresponding to 13 OTU sequences. The lack of match between environmental sequences and culture-based methods is a general trend, already mentioned by several articles on endophytes (e.g., Sun and Guo, 2012; Singh et al., 2017) and probably amplified by the fact that we conducted culturing of endophytes from two batches of samples, but NGS amplification from only one of these, albeit from the same plants and leaves in the forest. When exploring the co-occurrence values of nine cultures which displayed antipathogenic activity, it was found that within the whole endophyte community, these exhibited neither strong aggregative nor exclusionary effects, indicating that detected antipathogen activity under culture did not necessarily translate to determining community composition within leaves. Overall however, when cultures were bacterial, they tended to co-occur to a lesser extent with bacteria, and when cultures were fungal, they tended to co-occur to a lesser extent with fungi. Although this demonstrates that the cultured endophytes which display antipathogenic activity do not elicit an effect in a mature leaf, it does not necessarily imply that they would not prevent the arrival of a future pathogen. Tests to this effect have been conducted by numerous authors to detect the effects of tropical endophytes in conferring protection to their hosts, using seedlings which have been grown in absence of endophytes, such as Theobroma cacao seedlings and the pathogenic fungi Phytophthora (Arnold et al., 2003; Herre et al., 2007).
The work in this study does not support the idea of a community wide endophyte response to pathogens, but instead indicates that specific OTUs are likely to demonstrate an antipathogenic role. Given the long lived nature of palm leaves, such trends in endophyte dynamics may not reflect what happens in trees in deciduous forests which lose their leaves each year. Similarly, the response of endophyte communities in forests with high pathogen pressure, such as a habitat under environmental stress could prove markedly different from those recorded here. The idea of a microbiome conferring protection against pathogens has also been explored in animal systems, for example amphibian cuticular bacterial communities and their response to the fungus Batrachochytrium dendrobatidis (Hughey et al., 2017; Catenazzi et al., 2018), which like this study, found that whilst whole community composition did not shift in response to pathogen presence, specific OTUs did.
Although the above approach has demonstrated potential in revealing patterns of co-occurrence and competition, it can be considered as having several flaws. Firstly, the two pathogens used in the study, whilst common in laboratory tests of activity, are not common pathogens in the forests of French Guiana. An alternative approach to testing activity could be the use of co-culture techniques between isolated endophytes with known pathogens, such as Pycnoporus sanguineus, a Polyporaceae which is responsible for plant damage in French Guiana and the tropics and used to detect antipathogen activities (Rodrigues et al., 2010). Similarly, co-culturing of endophytes collected from the same organism in culture would reveal the direct interaction between species without the influence of other members of the community (Casella et al., 2013). Co-culturing tests have revealed a wealth of information as to the potential of co-occurring microorganisms to produce bioactive compounds of interest to biochemists, famously the accidental discovery of Penicillin (Fleming, 1929), but also the endophyte derived chemotherapy drug Taxol (Strobel et al., 1993). However, they are less successful in mirroring the interactions which are likely to occur in natural systems, since they rely on the supposition of one-way interactions between two species, when in reality, interactions within a community of endophytes are likely to be much more complex.
The above analysis has nonetheless proven valuable in demonstrating antipathogenic activity in a select number of cultures which were directly harvested from a plant host. It has shown that a majority of endophytes do not appear to be competitive, and that only a minority are active against pathogens. In spite of an absence of detected antipathogenic activity conferred by the endophyte community, the sampled palms were apparently in full health. Although this could point to plant foliar traits such as leaf structure or chemistry as a means to avoid pathogen establishment (e.g., Sanchez-Azofeifa et al., 2012), it does not necessarily mean that the detected endophytes play no functional role. In order for palms to have such long lifespans, endophytes could potentially confer greater resistance to drought, or as a defense against phytophagy by invertebrates. Finally, beyond their use as a means to detect pathogen activity, foliar endophytes represent a unique system to explore the effects of competition for resources due to the high numbers of species occurring within a habitat of defined boundaries.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JD, MB, VE, DS, and MR contributed to the conception and design of the study. YE, VE, DS, and MR conducted field collection of specimens. SM and MR performed NGS based methods. JD and MR performed the bioinformatic processing of the sequence data and wrote the first draft of the manuscript. MB and VE performed culturing and activity assays organized the database. JD and NG performed the statistical analysis. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by a joint Agence Nationale de la Recherche-Swiss National Fund (ANR-SNF) grant (SECIL, ref. ANR-15-CE21-0016 and SNF N° 310030E-164289) and from an Investissement d'Avenir grants also managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-25-01; ANAEE-France: ANR-11-INBS-0001, TULIP: ANR-10-LABX-0041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Emeline Houël and Guillaume Odonne for their help in the field, the BIO2MAR platform and Laurent Interaglia (http://bio2mar.obs-banyuls.fr) for performing ITS and 16S sequencing on isolated strains. We are grateful to the Genotoul bioinformatics platform (Toulouse, Midi-Pyrénées) for providing computing resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2019.00086/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alvarez-Sanchez, J., Barajas-Guzman, G., Campo, J., and Leon, R. (2016). Inorganic nitrogen and phosphorus in stemflow of the palm Astrocaryum mexicanum liebm. located in Los Tuxtlas, Mexico. Trop. Ecol. 57, 45–55.
Alvarez-Sanchez, J., and Guevara, S. (1999). Litter interception on Astrocaryum mexicanum liebm. (Palmae) in a tropical rain forest. Biotropica 31, 89–92. doi: 10.1111/j.1744-7429.1999.tb00119.x
André, A., Wojtowicz, N., Touré, K., Stien, D., and Eparvier, V. (2017). New acorane sesquiterpenes isolated from the endophytic fungus Colletotrichum gloeosporioides SNB-GSS07. Tetrahedron Lett. 58, 1269–1272. doi: 10.1016/j.tetlet.2017.02.024
Arnold, A. E., and Engelbrecht, B. M. (2007). Fungal endophytes nearly double minimum leaf conductance in seedlings of a neotropical tree species. J. Trop. Ecol. 23, 369–372. doi: 10.1017/S0266467407004038
Arnold, A. E., and Herre, E. A. (2003). Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 95, 388–398. doi: 10.1080/15572536.2004.11833083
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Arnold, A. E., Maynard, Z., Gilbert, G. S., Coley, P. D., and Kursar, T. A. (2000). Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3, 267–274. doi: 10.1046/j.1461-0248.2000.00159.x
Arnold, A. E., Mejía, L. C., Kyllo, D., Rojas, E. I., Maynard, Z., Robbins, N., et al. (2003). Fungal endophytes limit pathogen damage in a tropical tree. PNAS 100, 15649–15654. doi: 10.1073/pnas.2533483100
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6:343. doi: 10.1038/ismej.2011.119
Beckers, B., De Beeck, M. O., Thijs, S., Truyens, S., Weyens, N., Boerjan, W., et al. (2016). Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Fron. Microbiol. 7:650. doi: 10.3389/fmicb.2016.00650
Bertrand, S., Bohni, N., Schnee, S., Schumpp, O., Gindro, K., and Wolfender, J. (2014). Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 32, 1180–1204. doi: 10.1016/j.biotechadv.2014.03.001
Bertrand, S., Schumpp, O., Bohni, N., Monod, M., Gindro, K., and Wolfender, J. (2013). De novo production of metabolites by fungal co-culture of trichophyton rubrum and Bionectria ochroleuca. J. Nat. Prod. 76, 1157–1165 doi: 10.1021/np400258f
Bittleston, L. S., Brockmann, F., Wcislo, W., and Van Bael, S. A. (2010). Endophytic fungi reduce leaf-cutting ant damage to seedlings. Biol. Lett. 7, 30–32. doi: 10.1098/rsbl.2010.0456
Bohan, D. A., Vacher, C., Tamaddoni-Nezhad, A., Raybould, A., Dumbrell, A. J., and Woodward, G. (2017). Next-generation global biomonitoring: Large-scale, automated reconstruction of ecological networks. Trends Ecol. Evol. 32, 477–487. doi: 10.1016/j.tree.2017.03.001
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). Obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi: 10.1111/1755-0998.12428
Cannon, P. F., and Simmons, C. M. (2002). Diversity and host preference of leaf endophytic fungi in the Iwokrama Forest Reserve, Guyana. Mycologia 94, 210–220. doi: 10.1080/15572536.2003.11833226
Carroll, G. (1988). Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology 69, 2–9. doi: 10.2307/1943154
Casella, T. M., Eparvier, V., Mandavid, H., Bendelac, A., Odonne, G., Dayan, L., et al. (2013). Antimicrobial and cytotoxic secondary metabolites from tropical leaf endophytes: Isolation of antibacterial agent pyrrocidine C from Lewia infectoria SNB-GTC2402. Phytochemistry 96, 370–377. doi: 10.1016/j.phytochem.2013.10.004
Catenazzi, A., Flechas, S. V., Burkart, D., Hooven, N. D., Townsend, J., and Vredenburg, V. T. (2018). Widespread elevational occurrence of antifungal bacteria in Andean amphibians decimated by disease: a complex role for skin symbionts in defense against chytridiomycosis. Front. Microbiol. 9:465. doi: 10.3389/fmicb.2018.00465
Charles-Dominique, P., Chave, J., Dubois, M., De Granville, J., Riera, B., and Vezzoli, C. (2003). Colonization front of the understorey palm Astrocaryum sciophilum in a pristine rain forest of French Guiana. Glob. Ecol. Biogeogr.12, 237–248. doi: 10.1046/j.1466-822X.2003.00020.x
Charles-Dominique, P., Chave, J., Vezzoli, C., Dubois, M.-A., and Riéra, B. (2001). “Growth strategy of the understorey palm Astrocaryum sciophilum in the rainforest of French Guiana,” in Life Forms and Dynamics in Tropical Forests, eds G. Gottsberger and S. Liede (Berlin; Stuttgart), 153–163.
Chase, J. M., and Leibold, M. A. (2003). Ecological Niches: Linking Classical and Contemporary Approaches. Chicago, IL: University of Chicago Press.
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Christian, N., Herre, E. A., and Clay, K. (2019). Foliar endophytic fungi alter patterns of nitrogen uptake and distribution in Theobroma cacao. New Phytol. 222, 1573–1583. doi: 10.1111/nph.15693
Coissac, E., Riaz, T., and Puillandre, N. (2012). Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 21, 1834–1847. doi: 10.1111/j.1365-294X.2012.05550.x
Cordier, T., Robin, C., Capdevielle, X., Fabreguettes, O., Desprez-Loustau, M. L., and Vacher, C. (2012). The composition of phyllosphere fungal assemblages of European beech (Fagus sylvatica) varies significantly along an elevation gradient. New Phytol. 196, 510–519. doi: 10.1111/j.1469-8137.2012.04284.x
Crowther, T. W., Boddy, L., and Maynard, D. S. (2018). The use of artificial media in fungal ecology. Fung. Ecol. 32, 87–91. doi: 10.1016/j.funeco.2017.10.007
Dormann, C. F., Gruber, B., and Fründ, J. (2008). Introducing the bipartite package: analysing ecological networks. Interaction 8, 8–11.
Epp, L. S., Boessenkool, S., Bellemain, E. P., Haile, J., Esposito, A., Riaz, T., et al. (2012). New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Mol. Ecol. 21, 1821–1833. doi: 10.1111/j.1365-294X.2012.05537.x
European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2016). Available online at: http://www.eucast.org (accessed April 11, 2016).
Fagervold, S. K., Urios, L., Intertaglia, L., Batailler, N., Lebaron, P., and Suzuki, M. T. (2013). Pleionea mediterranea gen. nov., sp. nov., a gammaproteobacterium isolated from coastal seawater. Int. J. System. Evol. Microbiol. 63, 2700–2705. doi: 10.1099/ijs.0.045575-0
Ficetola, G. F., Coissac, E., Zundel, S., Riaz, T., Shehzad, W., Bessière, J., et al. (2010). An in silico approach for the evaluation of DNA barcodes. BMC Genom. 11:434. doi: 10.1186/1471-2164-11-434
Fleming, A. (1929). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10:226.
Fliegerova, K., Tapio, I., Bonin, A., Mrazek, J., Callegari, M. L., Bani, P., et al. (2014). Effect of DNA extraction and sample preservation method on rumen bacterial population. Anaerobe 29, 80–84. doi: 10.1016/j.anaerobe.2013.09.015
Fróhlich, J., Hyde, K., and Petrini, O. (2000). Endophytic fungi associated with palms. Mycol. Res. 104, 1202–1212. doi: 10.1017/S095375620000263X
Gazis, R., and Chaverri, P. (2015). Wild trees in the Amazon basin harbor a great diversity of beneficial endosymbiotic fungi: is this evidence of protective mutualism? Fungal Ecol. 17, 18–29. doi: 10.1016/j.funeco.2015.04.001
Gotelli, N. J. (2000). Null model analysis of species co-occurrence patterns. Ecology 81, 2606–2621. doi: 10.1890/0012-9658(2000)081[2606:NMAOSC]2.0.CO;2
Gotelli, N. J., and Ellison, A. M. (2013). EcoSimR 1.00. Available online at: http://www.uvm.edu/~ngotelli/EcoSim/EcoSim.html
Griffin, E. A., and Carson, W. P. (2015). The ecology and natural history of foliar bacteria with a focus on tropical forests and agroecosystems. Bot. Rev. 81, 105–149. doi: 10.1007/s12229-015-9151-9
Griffin, E. A., and Carson, W. P. (2018). “Tree endophytes: cryptic drivers of tropical forest diversity,” in Endophytes of Forest Trees: Biology and Applications, eds A. M. Pirttilä, and C. Frank (Basel Springer), 63–103.
Griffin, E. A., Traw, M. B., Morin, P. J., Pruitt, J. N., Wright, S. J., and Carson, W. P. (2016). Foliar bacteria and soil fertility mediate seedling performance: a new and cryptic dimension of niche differentiation. Ecology 97, 2998–3008. doi: 10.1002/ecy.1537
Haruna, E., Zin, N. M., Kerfahi, D., and Adams, J. M. (2018). Extensive overlap of tropical rainforest bacterial endophytes between soil, plant parts, and plant species. Microb. Ecol. 75, 88–103. doi: 10.1007/s00248-017-1002-2
Herre, E. A., Mejía, L. C., Kyllo, D. A., Rojas, E., Maynard, Z., Butler, A., et al. (2007). Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology 3, 550–558. doi: 10.1890/05-1606
Higgins, K. L., Arnold, A. E., Coley, P. D., and Kursar, T. A. (2014). Communities of fungal endophytes in tropical forest grasses: highly diverse host-and habitat generalists characterized by strong spatial structure. Fungal Ecol. 8, 1–11. doi: 10.1016/j.funeco.2013.12.005
Hsieh, T. C., Ma, K. H., and Chao, A. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol Evol. 7, 1451–1456. doi: 10.1111/2041-210X.12613
Hughey, M. C., Pena, J. A., Reyes, R., Medina, D., Belden, L. K., and Burrowes, P. A. (2017). Skin bacterial microbiome of a generalist Puerto Rican frog varies along elevation and land use gradients. PeerJ 5:e3688. doi: 10.7717/peerj.3688
Izuno, A., Kanzaki, M., Artchawakom, T., Wachrinrat, C., and Isagi, Y. (2016). Vertical structure of phyllosphere fungal communities in a tropical forest in thailand uncovered by high-throughput sequencing. PLoS ONE 11:e0166669. doi: 10.1371/journal.pone.0166669
Jakuschkin, B., Fievet, V., Schwaller, L., Fort, T., Robin, C., and Vacher, C. (2016). Deciphering the pathobiome: intra-and interkingdom interactions involving the pathogen Erysiphe alphitoides. Microb. Ecol. 72, 870–880. doi: 10.1007/s00248-016-0777-x
Kennedy, P. G., Peay, K. G., and Bruns, T. D. (2009). Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology 90, 2098–2107. doi: 10.1890/08-1291.1
Kennedy, P. G., Weber, M. G., and Bluhm, A. A. (2010). Frankia bacteria in Alnus rubra forests: genetic diversity and determinants of assemblage structure. Plant Soil 335, 479–492. doi: 10.1007/s11104-010-0436-9
Maignien, L., DeForce, E. A., Chafee, M. E., Eren, A. M., and Simmons, S. L. (2014). Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 5, e00682–e00613 doi: 10.1128/mBio.00682-13
Nirma, C., Eparvier, V., and Stien, D. (2013). Antifungal agents from Pseudallescheria boydii SNB-CN73 isolated from a Nasutitermes sp. termite. J. Nat. Prod. 76, 988–991. doi: 10.1021/np4001703
Quast, C., Pruesse, E., Yilmaz, P., Gerken, P., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rodrigues, A. M. S., Eparvier, V., Odonne, G., Amusant, N., Stien, D., and Houël, E. (2019). The antifungal potential of (Z)-ligustilide and the protective effect of eugenol demonstrated by a chemometric approach. Sci. Rep. 9:8729. doi: 10.1038/s41598-019-45222-y
Rodrigues, A. M. S., Theodoro, P. N. E. T., Eparvier, V., Basset, C., Silva, M. R. R., Beauchêne, J., et al. (2010). Search for antifungal compounds from the wood of durable tropical trees. J. Nat. Prod. 73, 1706–1707. doi: 10.1021/np1001412
Rodrigues, K. F. (1994). The foliar fungal endophytes of the Amazonian palm Euterpe oleracea. Mycologia 86, 376–385. doi: 10.1080/00275514.1994.12026423
Saikkonen, K., Faeth, S. H., Helander, M., and Sullivan, T. J. (1998). Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 29, 319–343. doi: 10.1146/annurev.ecolsys.29.1.319
Sanchez-Azofeifa, A., Oki, Y., Fernandes, G. W., Ball, R. A., and Gamon, J. (2012). Relationships between endophyte diversity and leaf optical properties. Trees 26, 291–299. doi: 10.1007/s00468-011-0591-5
Singh, D. K., Sharma, V. K., Kumar, J., Mishra, A., Verma, S. K., Sieber, T. N., et al. (2017). Diversity of endophytic mycobiota of tropical tree Tectona grandis Linn.f.: spatiotemporal and tissue type effects. Sci. Rep. 7:3745. doi: 10.1038/s41598-017-03933-0
Solis, M. J. L., Cruz, T. E. D., Schnittler, M., and Unterseher, M. (2016). The diverse community of leaf-inhabiting fungal endophytes from Philippine natural forests reflects phylogenetic patterns of their host plant species Ficus benjamina, F. elastica and F. religiosa. Mycoscience 57, 96–106. doi: 10.1016/j.myc.2015.10.002
Strobel, G. A., Stierle, A., and Hess, W. M. (1993). Taxol formation in yew—Taxus. Plant Sci. 92, 1–12. doi: 10.1016/0168-9452(93)90060-D
Sun, X., and Guo, L. (2012). Endophytic fungal diversity: review of traditional and molecular techniques. Mycology 3, 65–76.
Taberlet, P., Bonin, A., Zinger, L., and Coissac, E. (2018). Environmental DNA: For Biodiversity Research and Monitoring. Oxford, UK: Oxford University Press.
Truong, C., Mujic, A. B., Healy, R., Kuhar, F., Furci, G., Torres, D., et al. (2017). How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytol. 214, 913–919. doi: 10.1111/nph.14509
Vacher, C., Hampe, A., Porté, A. J., Sauer, U., Compant, S., and Morris, C. E. (2016). The phyllosphere: microbial jungle at the plant–climate interface. Annu. Rev. Ecol. Evol. Syst. 47, 1–24. doi: 10.1146/annurev-ecolsys-121415-032238
Van Bael, S., Estrada, C., and Arnold, A. E. (2017). “Chapter 6: foliar endophyte communities and leaf traits in tropical trees,” in The Fungal Community: Its Organization and Role in the Ecosystem, eds J. Dighton and J. F. White (Boca Raton, FL: CRC Press, 79–94.
Vincent, J. B., Weiblen, G. D., and May, G. (2016). Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol. Ecol. 25, 825–841. doi: 10.1111/mec.13510
Weber, R. W., Stenger, E., Meffert, A., and Hahn, M. (2004). Brefeldin A production by Phoma medicaginis in dead pre-colonized plant tissue: a strategy for habitat conquest? Mycol. Res. 108, 662–671. doi: 10.1017/S0953756204000243
Wei, T., Simko, V., Levy, M., Xie, Y., Jin, Y., and Zemla, J. (2017). Package ‘corrplot'. Statistician 56, 316–324.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Williams, R. J., Howe, A., and Hofmockel, K. S. (2014). Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front. Microbiol. 5:358 doi: 10.3389/fmicb.2014.00358
Zimmerman, N. B., and Vitousek, P. M. (2012). Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. PNAS 109, 13022–13027. doi: 10.1073/pnas.1209872109
Keywords: endophyte, fungi, bacteria, pathogen, rainforest, cultures, palm, metabarcoding
Citation: Donald J, Barthélemy M, Gazal N, Eveno Y, Manzi S, Eparvier V, Stien D and Roy M (2019) Tropical Palm Endophytes Exhibit Low Competitive Structuring When Assessed Using Co-occurrence and Antipathogen Activity Analysis. Front. For. Glob. Change 2:86. doi: 10.3389/ffgc.2019.00086
Received: 08 August 2019; Accepted: 03 December 2019;
Published: 18 December 2019.
Edited by:
Julieta Benitez-Malvido, National Autonomous University of Mexico, MexicoReviewed by:
Jonàs Oliva, Universitat de Lleida, SpainPeter Matthew Scott, The New Zealand Institute for Plant & Food Research Ltd., New Zealand
Copyright © 2019 Donald, Barthélemy, Gazal, Eveno, Manzi, Eparvier, Stien and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian Donald, julesdonald@gmail.com
 Julian Donald
Julian Donald Morgane Barthélemy2
Morgane Barthélemy2  Nina Gazal
Nina Gazal Didier Stien
Didier Stien Mélanie Roy
Mélanie Roy