- 1Faculty of Science, Kunming University of Science and Technology, Kunming, China
- 2Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, China
Large-scale intensive cultivation has made continuous cropping soil sickness more serious for Panax notoginseng in Yunnan. Autotoxic substances can promote the occurrence of continuous cropping soil sickness. Phenolic acids exert a strong autotoxic effect on P. notoginseng. Based on UPLC-MS/MS, the levels of six phenolic acids with the strongest autotoxicity of P. notoginseng rhizospheric soil were tested. Based on Illumina MiSeq high-throughput sequencing technology, the variation in the microbial diversity in the rhizospheric soil was used as an index to explore the interactions between phenolic acids and the soil microorganisms of the P. notoginseng rhizosphere. (1) Continuous P. notoginseng cropping significantly changed the microbial community structure. Continuous cropping increased bacterial Chao1 index and Shannon index and decreased fungal Shannon index. After P. notoginseng disease, bacterial Shannon index reduced and fungal Chao1 index decreased. (2) Phenolic acid significantly changed the bacterial community structure. VA significantly reduced the bacterial Shannon index. Exogenous p-HA, FA, SA, and VA significantly increased the fungal Chao1 index and p-HA showed the most significant effect. Para-HA affected bacterial specificity, and VA affected fungal specificity. (3) VA was positively correlated with most fungi and bacteria. Para-HA was positively correlated with Lelliottia and Flavobacterium. Para-HA was also positively correlated with plant pathogens (Fusarium and Ilyonectria). Para-HA and VA were able to promote the growth of primary pathogenic bacteria. Thus, p-HA and VA are the main phenolic acid-autotoxin substances in P. notoginseng under continuous cropping. (4) A correlation analysis of soil environmental factors associated with fungal and bacterial communities showed that AK, TN, OM, and HN were most strongly correlated with soil microorganisms. (5) The microorganisms in the rhizosphere of 3-year-old soil planted with P. notoginseng exhibited obvious effects on the degradation of the four phenolic acids. The effect of soil microorganisms on phenolic acids was first-order kinetic degradation with a high degradation rate and a half-life of less than 4.5 h. The results showed that phenolic acids could promote the growth of pathogenic bacteria. And the interaction between rhizospheric soil microorganisms and phenolic acids was the main cause of the disturbance of P. notoginseng rhizosphere microflora.
Introduction
Sanqi [Panax notoginseng (Burkill) F. H. Chen] is a perennial medicinal plant in the genus Panax. The dried rhizomes of P. notoginseng can relieve blood stasis and stop bleeding, promote blood circulation and relieve pain. These rhizomes are primarily used to treat hemorrhagic disease, bruising injuries, blood stasis, swelling, and pain (Macdermot, 1950; Commission, 2015; Xu et al., 2018). Wenshan Prefecture in Yunnan Province is an area that is famous for the production of genuine P. notoginseng; it has a warm and rainy climate that is very suitable for growing P. notoginseng. The growth cycle of P. notoginseng is generally 3 years or more, and the cultivation process requires large amounts of fertilizers and pesticides, which lead to the deterioration of the physical and chemical properties of the soil, an imbalance of nutrients and microbial communities, and allelopathy and autotoxicity; collectively, these ailments are commonly known as the continuous cropping soil sickness of P. notoginseng (Tan et al., 2017). Due to the increasing demand for P. notoginseng in recent years, large-scale intensive planting has made continuous cropping soil sickness increasingly serious, which strongly affects the quality and yield of P. notoginseng (Yang et al., 2016; Qiao et al., 2020).
There are many factors that lead to continuous cropping soil sickness, including root exudates that induce changes in rhizospheric soil microorganisms, allelopathic interactions of soilborne pathogens, imbalances of rhizospheric microflora aggravated soil acidification, aggravation of soil fungal diseases caused by increases in rhizospheric pathogens and declines in beneficial bacteria, and the development of viral diseases (Wu and Lin, 2020). Alteration of soil microbial community structure is one of the important reasons for continuous cropping soil sickness. The numbers of microorganisms and physiological groups as well as the levels of soil enzyme activities and nutrients in soil planted in P. notoginseng across different planting and fallow years were studied. The structure of the soil microbial community and the soil nutrient balance were found to have changed after P. notoginseng cultivation. It was assumed that the changes in the biological community were not conducive to P. notoginseng growth (Zhang et al., 2019). Other studies have also found that an increase in the number of P. notoginseng planting years will cause an imbalance in the rhizospheric microbiota, increase the abundance of pathogenic fungal genera in the rhizosphere, decrease the abundance of some beneficial fungal genera, weaken the growth tendency of P. notoginseng, and significantly increase the incidence of disease (Tang et al., 2020). Long-term continuous cropping has been found to reduce the soil microbial diversity of the cotton (Gossypium spp.) rhizosphere and resulted in continuous cropping soil sickness, which led to a reduced cotton yield (Gu et al., 2012). Continuous cropping of maize (Zea mays Linn.) destroys the balance of the soil microflora, reducing the number of beneficial microbes and increasing populations of soil fungi, which causes serious disease and reduces the main soil nutrients (Xing et al., 2011). Continuous cucumber cropping significantly affects the abundance and community structure of soil fungi and increases the incidence of soil diseases (Li Y. et al., 2020). Therefore, changes in the rhizospheric soil microbial community can affect the growth of host plants.
Studies have shown that during the continuous cropping process, plants primarily communicate with the soil through root exudates, which can directly or indirectly change the soil microbial community, thus promoting the occurrence of continuous cropping soil sickness (Wu and Lin, 2020). For example, exudates of false starwort (Pseudostellaria heterophylla) roots have been found to mediate changes in soil microbial community structure and functional diversity, which affected the growth of the plants (Wu et al., 2016). Peanut (Arachis hypogaea Linn.) root exudates selectively inhibit or stimulate certain bacterial and fungal groups, promote changes in the soil microbial community and inhibit the growth of peanut plants (Li Y. et al., 2014). In conclusion, interactions between chemical substances secreted by plant roots and rhizospheric soil microorganisms cause changes in the rhizospheric soil microbial community, which is the primary reason for continuous cropping soil sickness (Yang et al., 2012; Dong et al., 2016; Qian et al., 2016; Tan et al., 2017; Feng et al., 2020).
Chemicals in rhizospheric soil influence microbial community composition. Allelochemicals can reduce soil microbial diversity, significantly affect the genetic structure and carbon metabolism capacity of microbial communities, and lead to imbalances in soil microbial ecosystems (Li X. et al., 2014). Some researchers have simulated the selective effects of allelochemicals secreted by plant roots on different microorganisms in the rhizosphere, and have found that root exudates can selectively inhibit the growth of beneficial microorganisms and promote the proliferation of pathogenic microorganisms in the soil (Wu et al., 2016). Metabolites of P. notoginseng are detected in rhizospheric soil allelochemicals and primarily include p-hydroxybenzoic acid, vanillic acid, syringic acid, p-coumaric acid, ferulic acid, and benzoic acid. Allelopathy testing has confirmed their order from strong to weak: p-coumaric acid, benzoic acid, vanillic acid, ferulic acid, p-hydroxybenzoic acid, and syringic acid (Wu et al., 2014). In a study on the influence of exogenous p-hydroxybenzoic acid on the soil microbial community of the cucumber rhizosphere, p-hydroxybenzoic acid decreased the Shannon–Wiener index of the rhizospheric bacterial community and increased the Shannon–Wiener index of the rhizospheric fungal community (Zhou et al., 2012). Different concentrations of cinnamic acid, phthalic acid, p-hydroxybenzoic acid and their mixtures have been found to decrease the quantities of peanut rhizospheric soil bacteria and actinomycetes, microbial biomass carbon and nitrogen content, and respiration intensity, increase the quantity of fungi, and promote the occurrence of soil sickness in peanut (Liu et al., 2018). Studies have also shown that phenolic acids such as gallic acid, salicylic acid, hydrocinnamic acid, and benzoic acid can increase fungal richness, change fungal community structure and increase pathogen loads (Li et al., 2018). Previous studies have confirmed that phenolic acids can significantly affect the biomass, diversity and community structure of soil microorganisms (Qu and Wang, 2008).
Several studies have shown that the accumulation of phenolic acids secreted by plant roots can mediate changes in the structure and functional diversity of soil microbial communities and indirectly affect the growth and development of host plants (Wu et al., 2016, 2017). Therefore, it has been speculated that interactions between phenolic acids secreted by P. notoginseng roots and rhizospheric soil microorganisms may disrupt the root microecological balance and promote the occurrence of continuous cropping soil sickness. However, there are still no reports on this topic. An increasing number of studies have shown that disturbances of the microflora mediated by root exudates play a key role in continuous cropping soil sickness (Wu et al., 2016). However, studies on continuous cropping soil sickness of P. notoginseng primarily focus on the isolation, identification and biological testing of allelochemicals, and the regulation of soil microorganisms by phenolic acids during P. notoginseng planting remains relatively unclear. On this basis, the interactions between phenolic acids secreted by P. notoginseng roots and soil microorganisms were studied here.
The innovative aspect of this study lies in its application of results from both field investigations and laboratory experiments to determine the interactions between phenolic acids and P. notoginseng rhizospheric soil microorganisms. To simulate P. notoginseng continuous cropping, autotoxic substances were added exogenously to regulate rhizospheric soil microorganisms. In addition, microorganisms exhibit detoxification functions and can degrade autotoxins in soil. This study systematically and comprehensively revealed the interactions between autotoxins and the rhizospheric soil microbial-plant cycle. Here, healthy and diseased rhizospheric soils under P. notoginseng cultivation for different numbers of years in Wenshan Prefecture, Yunnan Province were selected. UPLC-MS/MS and Illumina MiSeq high-throughput sequencing technology were used to determine the contents of benzoic acid (BA), p-hydroxybenzoic acid (p-HA), ferulic acid (FA), syringic acid (SA), vanillic acid (VA), and p-coumaric acid (p-CA) and microbial community structure in different soils (Wu et al., 2014). Interactions between phenolic acid autotoxic substances and soil microorganisms from the P. notoginseng root system were explored to provide a basis for solving these obstacles to continuous cropping. The study was designed to test these hypotheses: (1) The concentration of phenolic acids in P. notoginseng rhizospheric soil is related to the health status and number of years of P. notoginseng planting. (2) Continuous cropping could significantly change the composition of the soil microbial community in the P. notoginseng rhizosphere. (3) Exogenous phenolic acids can change the composition of the soil microbial community. Phenolic acids are strongly associated with soil microorganisms and can promote the growth of plant pathogens. (4) Soil microorganisms can degrade phenolic acids and rapidly decrease their concentrations. Finally, the interaction between soil microorganisms and phenolic acids reached a dynamic balance. If balance is disturbed, disease can occur in host plants.
Materials and Methods
Site Description and Soil Sampling
In this study, rhizospheric soil planted with P. notoginseng for different numbers of years was collected to determine the phenolic acid contents of the soil. We analyzed the physical and chemical properties of soil samples and explored the interactions between phenolic acids and rhizospheric soil microorganisms. Soil samples were collected in Wenshan Prefecture, Yunnan Province (103.58′ N, 23.87′ E) in November 2020, and the sampling site was located near Pingyuan Street, Yanshan County. This area has a subtropical continental monsoon climate, with an annual average temperature of 25°C and an annual precipitation of 1,308 mm.
At the P. notoginseng cultivation site, control soil (CK), 1-year-old healthy rhizospheric soil (1Y), 2-year-old healthy rhizospheric soil (H-2Y), 2-year-old diseased rhizospheric soil (S-2Y), 3-year-old healthy rhizospheric soil (H-3Y), and 3-year-old diseased rhizospheric soil (S-3Y) were collected. Soil collected under the same conditions from 3 to 5 cm below the surface layer where nothing was planted was used as the control soil. Healthy rhizospheric soil refers to the soil attached to the root tubers of P. notoginseng plants that have been continuously planted for 2 or 3 years without disease or insect pests. The diseased soil was rhizospheric soil that from plants showing root rot from the same experimental plot (Tan et al., 2017). In this study, a total of six types of soils (CK, 1Y, H-2Y, S-2Y, H-3Y, and S-3Y) were collected. Three greenhouses were selected for each soil (the soil in each greenhouse was managed in the same way) for repeated collection using the five-point sampling method. The soil attached to the rhizosphere of P. notoginseng plants was shaken off into self-sealed bags, marked and brought back to the laboratory (Riley and Barber, 1970). Three biological replicates were performed for each type of soil sample. There were a total of 18 replicates among 6 types of soil sample. A portion of the soil was used for physicochemical property analysis and phenolic acid degradation experiments, while the remainder was stored at −80°C for subsequent soil microbial community diversity analysis.
Soil Physical and Chemical Properties
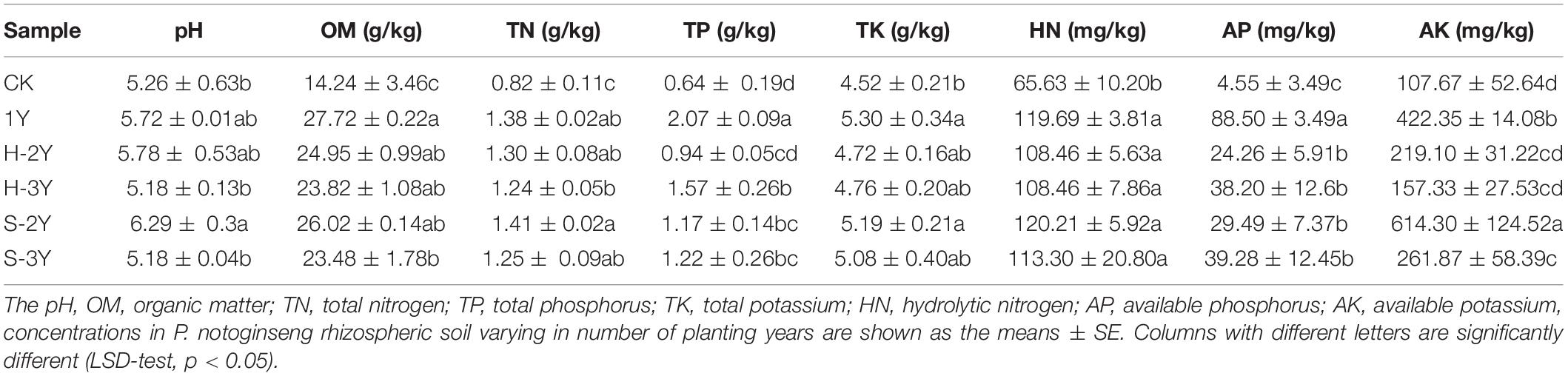
Control soil (CK), 1-year-old healthy rhizospheric soil (1Y), 2-year-old healthy (H-2Y) and diseased rhizospheric soil (S-2Y), and 3-year healthy (H-3Y) and diseased rhizospheric soil (S-3Y) were selected, and analysis of each sample was repeated five times to determine its physical and chemical properties. Soil pH was measured in a 1:2.5 soil-water (W/V) suspension with a pH 700 pH meter (United States, Uutech) (Qiu et al., 2012). The carbon dioxide content was quantitatively determined by potassium dichromate oxidation and external heating methods, and the organic matter (OM) content of each sample was calculated by carbon nitridation (Nelson and Sommers, 1996). The total nitrogen (TN) was determined using an elemental analyzer (Vario EL III, Germany) (Bremner, 1960). After each sample was infused with sodium hydroxide, the frit was dissolved in water and dilute sulfuric acid, the anti-chromogenic agents molybdenum and antimony were added to the solution, and the total phosphorus (TP) content in the soil was quantitatively determined with an ultraviolet spectrophotometer (L5) (GB/T9837-1988). The soil organic matter was removed by heating and oxidation with nitric acid and perchloric acid. After the silicate was digested with hydrofluoric acid, the remaining residue was dissolved with hydrochloric acid, and the total potassium (TK) in the soil was determined using a ZEENIT700P graphite furnace atomic absorption spectrometer (GB/T9836-1988). The hydrolytic nitrogen (HN) in the soil was determined by the alkaline hydrolysis diffusion method (Shen et al., 2011). The available phosphorus (AP) in the soil was extracted with sodium bicarbonate, and its content was determined by the molybdenum antimony anti-colorimetric method (Gyaneshwar et al., 2002). Extraction was performed with neutral ammonium acetate solution and soil available potassium (AK) was determined by flame photometry (Mclean and Watson, 1985). In this study, each sample was analyzed three times.
Determination of the Phenolic Acids Concentration in the Soil
Three g each of the control soil (CK), 1-year-old healthy rhizospheric soil (1Y), 2-year-old healthy rhizospheric soil (H-2Y), 2-year-old diseased rhizospheric soil (S-2Y), 3-year-old healthy rhizospheric soil (H-3Y), and 3-year-old diseased rhizospheric soil (S-3Y) were weighed and placed in separate 50 ml centrifuge tubes; then, 20 ml of chromatographic methanol was added to each tube, and phenolic acids were extracted ultrasonically for 40 min and centrifuged at 4,500 r/min for 10 min. After centrifugation, the residue remaining in each tube was incubated with 20 ml of methanol for ultrasonic extraction for 40 min and centrifugation at 4,500 r/min for 10 min (Dong et al., 2018). Each supernatant was combined twice and concentrated by rotary evaporation in a water bath at a temperature of 50°C until it was solvent-free. Then, 5 ml of methanol was added to each dried rotary steaming flask, and the solution was transferred to a 10 ml round-bottom flask after all the substances were dissolved. Each solution was concentrated to dry by rotary steaming (Zhong et al., 2020). The HLB columns were each washed with 2 ml methanol and then activated with 2 ml 1 M formic acid. Two milliliters of each extract (in a methanol:water:formic acid volumetric ratio of 15:4:1) was added to a rotary steaming flask, the supernatant was absorbed through the HLB column, and the eluent was collected. The HLB column was rinsed with 1 ml of extract, and the eluent was collected. The two eluents from each sample were combined for steam drying; then, 200 μl of extract was added and filtered through a 0.22 μm micron filter membrane. Quantitative analysis was performed using an UPLC-MS/MS (Tsushima 8050) in MRM mode (Jia et al., 2020). Each sample was analyzed three times.
Degradation of Phenolic Acids by Rhizospheric Soil Microorganisms
Generally, two and 3-year-old P. notoginseng showed the highest incidence of root rot. P. notoginseng planted in the field is generally harvested in 3 years. Considering the number of continuous cropping years and field practices used in P. notoginseng cultivation, 3-year-old healthy P. notoginseng rhizospheric soil was selected for the degradation experiment. Six samples of 36 g of 3-year-old healthy rhizospheric soil from around P. notoginseng plants was measured and placed in separate 250 ml tapered flasks with stoppers. Maximum values from results of the field investigation in this study and previous reports were adopted for the experimental setting. BA (5 μg/g), p-HA (5 μg/g), FA (5 μg/g), SA (5 μg/g), VA (10 μg/g), and p-CA (30 μg/g) (Wu et al., 2014) were each added to a separate conical flask containing a healthy soil sample and incubated in a thermostatic chamber at 25°C, and the conical flasks were aerated for 1 hour every 24 h. The soil moisture content was maintained at 70%. Three g of soil was randomly weighed from each conical flask at 0, 6, 12, 24, 48, and 72 h (Guo et al., 2011). The concentration of phenolic acids in each soil sample was quantitatively analyzed according to the first-order kinetic equations LnC(t)=LnC(t0)-Kt and t1/2=Ln2/K. The half-life of each autotoxic substance (t1/2) was calculated, where C is the concentration, T is the time, and K is the degradation rate (Xia et al., 2015). The method for determining soil phenolic acid concentrations was the same as that described above. Each phenolic acid sample was examined five times to explore the degradation ability of the soil microorganisms.
Total soil DNA was extracted from the rhizospheric soil 72 h after culture from 5 replicates and sequenced for analysis (Luo et al., 2020). The effects of the six phenolic acids on the soil microorganisms of P. notoginseng rhizospheres were studied.
DNA Extraction, PCR Amplification and Illumina MiSeq Sequencing
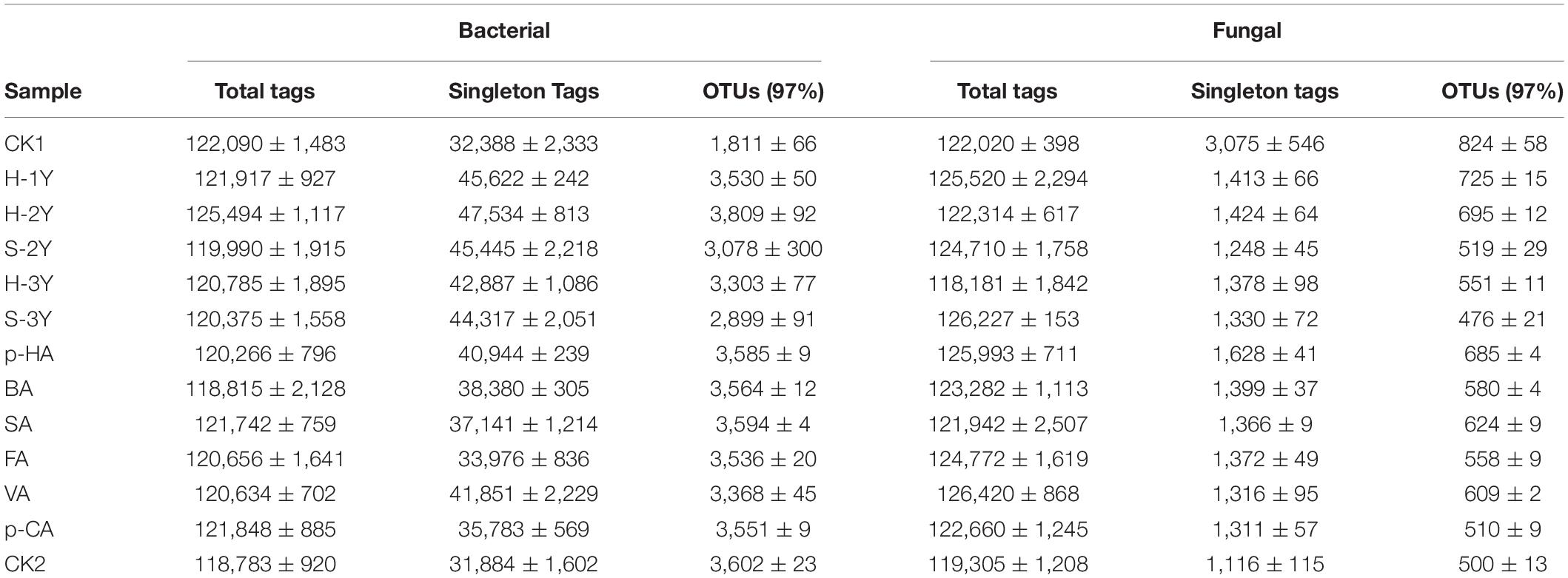
Total DNA from the soil microorganisms was extracted by using the DNeasy PowerSoil® Kit from QIAGEN, Germany. Three parallel samples were prepared for each sample and amplified by PCR (instrument model ETC811, Dongsheng Scientific Instruments Co., Ltd.). The diluted genomic DNA was used as a template and amplified with specific primers containing barcodes according to the selected sequencing region. The amplified products were subjected to gel electrophoresis, recovered and quantified using a QuantiFluor™ fluorometer. The bacterial genome (16S rDNA V3-V4 gene region) primers were 341F: CCTACGGGNGGCWGCAG and 806R: GGACTACHVGGGTA TCTAAT (Neefs et al., 1991). The fungal genome (ITS2 gene region) primers were ITS3-KYO2: GATGAAGAACGYA GYRAA and ITS4: TCCTCCGCTTATTGATATGC (Toju et al., 2012). Water was used as a negative control for the PCR procedure in each sample, and no bands were amplified from the negative controls. The purified amplification products were mixed in equal amounts, a sequencing connector was added to construct the sequencing libraries, and the Illumina PE250 library was sequenced on the machine. To ensure the reliability and validity of the data, FASTP was used to filter reads from the original dataset generated by the Illumina MiSeq platform. FASTP filtered out low-quality reads: (1) reads with over 10% of nucleotides (N) unknown were removed, and (2) reads for which less than 50% of bases showed quality scores (Q-values) > 20 were removed. FLASH was used to splice double-ended reads into tags and then filter low-quality tags to obtain clean tags (Magoc and Salzberg, 2011; Chen et al., 2018). The clean tags were clustered into operational taxonomic units (OTUs) of ≥ 97% similarity using the UPARSE (version 9.2.64) pipeline. All chimeric tags were removed using the UCHIME algorithm, and effective tags were finally obtained for further analysis. The tag sequence with the highest abundance was selected as the representative sequence within each cluster. All singleton sequencing data were removed from the sequencing data. OTUs were obtained by clustering with 97% sequence similarity (Table 1). The sequencing data were compared with SILVA (16S) and UNITE (ITS) database to obtain biotaxonomic information.
Data Analysis
We used QIIME software for alpha diversity index analysis (Kuczynski et al., 2012). Based on the intersample distance index, PCoA analysis was performed using the R language vegan package (Liu and Li, 2021). The degree and direction (positive or negative) of correlation between each environmental factor and each species were evaluated by Pearson analysis. FUNGuild was used to annotate fungal functions based on OTU (97%) abundance table information.
SPSS 21.0 software (SPSS Inc., United States) was used to analyze the data, and the homogeneity of variance was tested before the statistical analysis. Statistical analysis was performed using a one-way analysis of variance (ANOVA) and Duncan’s multi-interval test (P < 0.05). Sequencing data were provided by Guangzhou Gene Denovo Biotechnology Co., Ltd. Drawings and data analysis were completed by the first author of this study. Raw sequences were uploaded to the NCBI BioProject database (Accession: PRJNA757903).
Results
Analysis of Soil Physical and Chemical Properties
In the analysis of soil physical and chemical properties, the pH of the rhizospheric soil from planted P. notoginseng ranged from 5.18 to 6.29. The pH of the S-2Y rhizospheric soil around P. notoginseng plants was significantly higher than that of the control soil. Values of OM, TN, HN, and AP in healthy rhizospheric soil were significantly higher than those in the control soil. All the physical and chemical indexes of sick rhizospheric soil except pH were significantly higher than those of the control group. The AK content of healthy soil was significantly higher than that in sick rhizospheric soil (Table 2).
Determination and Analysis of Phenolic Acids in Rhizospheric Soil
The phenolic acids detected in P. notoginseng rhizospheric soil primarily included benzoic acid (BA), p-hydroxybenzoic acid (p-HA), ferulic acid (FA), syringic acid (SA), vanillic acid (VA), and p-coumaric acid (p-CA). The concentration of p-HA in the 2-year-old diseased rhizospheric soil (S-2Y) of P. notoginseng was significantly higher than that of the 3 years of rhizospheric soil (H-3Y and S-3Y). The concentration of SA in the 2-year-old diseased rhizospheric soil (S-2Y) of P. notoginseng was significantly higher than that of 1-year-old healthy rhizospheric soil (1Y), 2-year-old healthy rhizospheric soil (H-2Y) and 3-year-old diseased rhizospheric soil (S-3Y). The concentrations of p-HA (555.66 ± 121.95), FA (117.34 ± 9.86), and SA (58.28 ± 1.40) were highest in S-2Y among all the measured samples (Figure 1).
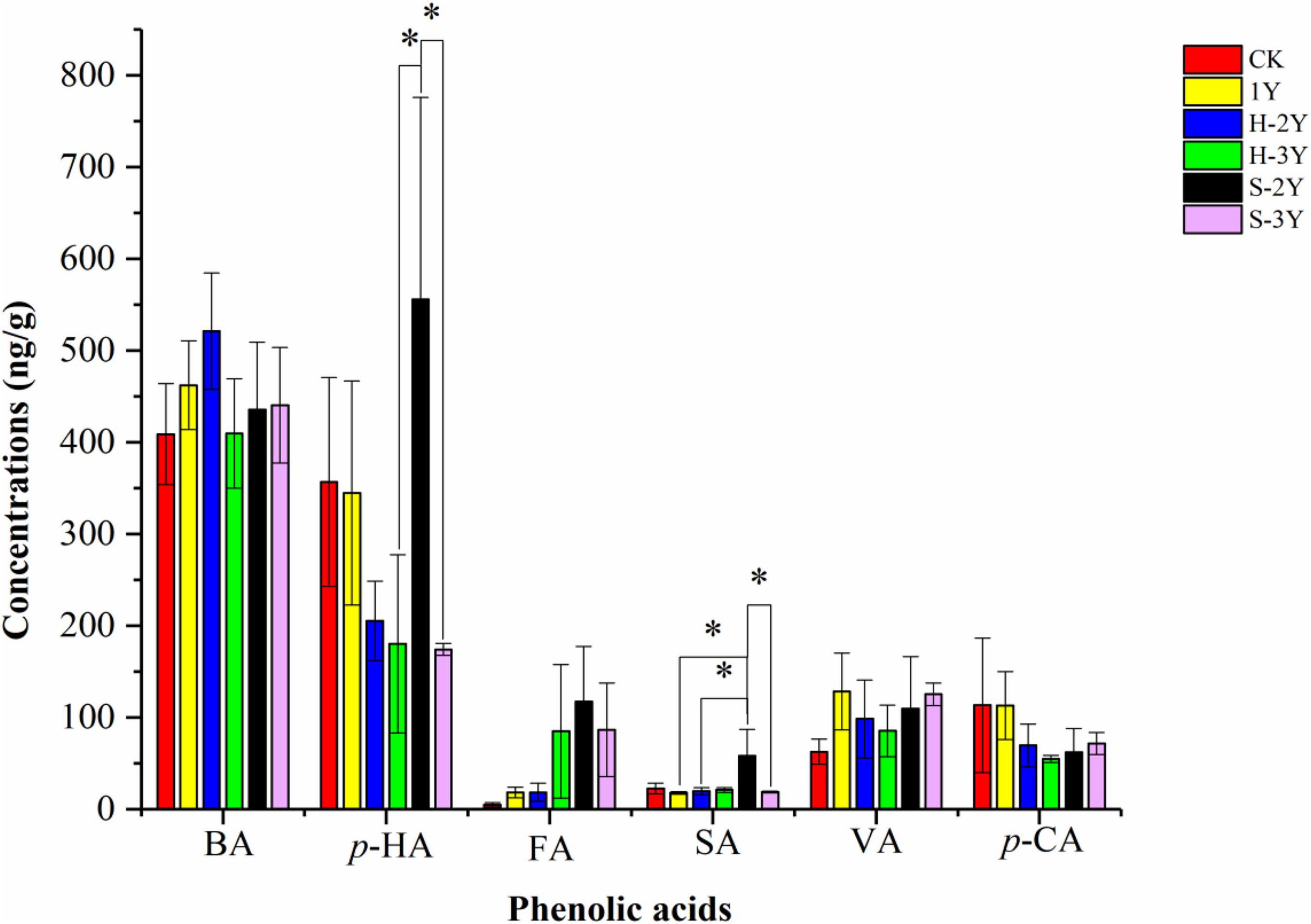
Figure 1. Phenolic acid concentrations. Red represents control soil, yellow represents 1-year-old P. notoginseng rhizospheric soil (H-1Y), blue represents 2-year-old healthy rhizospheric soil (H-2Y), green represents 3-year-old healthy rhizospheric soil (H-3Y), black represents 2-year-old diseased rhizospheric soil (S-2Y), and purple represents 3-year-old diseased rhizospheric soil (S-3Y). BA is benzoic acid, p-HA is p-hydroxybenzoic acid, FA is ferulic acid, SA is syringic acid, VA is vanillic acid, and p-CA is p-coumaric acid; * indicates a significant difference LSD-test, (p < 0.05).
Alpha Diversity Analysis
During the greenhouse experiment, bacterial Chao1 index in the rhizospheric soil from planted P. notoginseng were significantly higher than values for the control soil The Shannon index of bacteria in healthy P. notoginseng rhizosphere soil was significantly higher than that in control group. The Chao1 index of 3-year-old diseased P. notoginseng rhizospheric soil (S-3Y) was significantly lower than that of 2-year-old healthy P. notoginseng rhizospheric soil (H-2Y). The difference between Chao1 index and Shannon index may be caused by the uneven distribution of species. All the data indicated that continuous cropping led to an overall increase in bacterial Chao1 index and Shannon index, and the occurrence of disease reduced bacterial Shannon index. The exogenous addition of VA in the laboratory significantly reduced the bacterial Shannon index (Figure 2A).
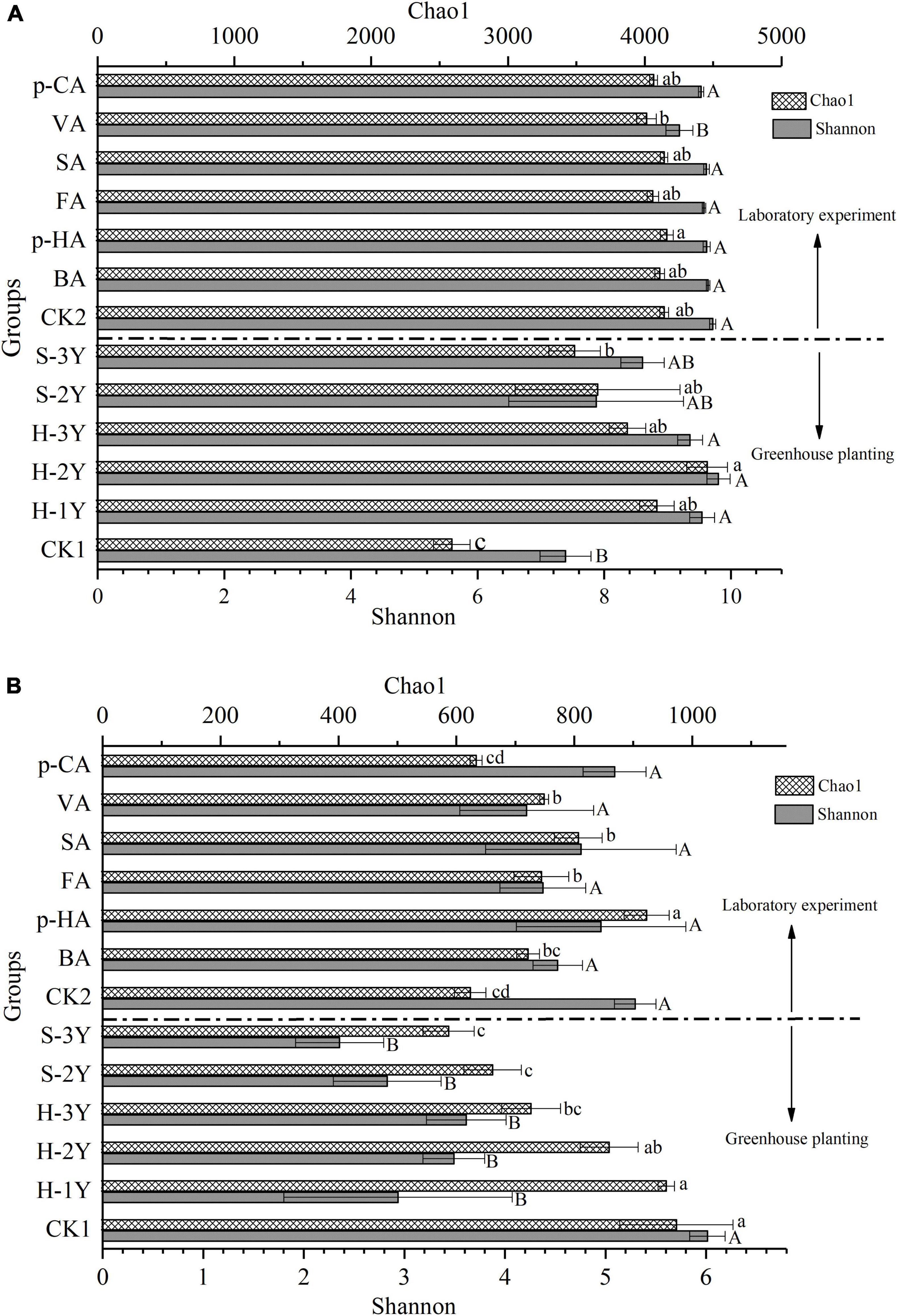
Figure 2. Changes in the rhizosphere soil microbial community associated with P. notoginseng. The bacterial Chao1 index and Shannon index (A) and fungal Chao1 index and Shannon index (B) are represented as the means ± SE (n = 3). CK1 is natural soil that has not been planted with P. notoginseng, and CK2 is the 3-year-old healthy rhizospheric soil from P. notoginseng cultivated for 72 h without exogenous phenolic acid addition. Different letters indicate significant differences (P < 0.05). Lowercase letters indicate the significant differences of Chao1 index, and uppercase letters indicate the significant differences of Shannon index. Above the dotted line are data from the indoor experiment, and below the dotted line are data from the greenhouse planting experiment.
Compared with the control soil, the rhizospheric soil planted with P. notoginseng significantly reduced fungal Shannon index in the greenhouse experiment. The fungal Chao1 index of diseased rhizospheric soil (S-2Y and S-3Y) was significantly lower than that of healthy rhizospheric soil (H-2Y and H-1Y) and control soil. The result showed that the fungal Chao1 index was significantly decreased after P. notoginseng disease. The Chao1 index for fungi in the rhizospheric soil decreased with increasing number of cultivation years, the 3-year-old healthy rhizosphere soil (H-3Y) decreased most significantly. Exogenous p-HA, FA, SA, and VA significantly increased the Chao1 index of the fungi compared with the control soil, and p-HA showed the most significant effect. Exogenous phenolic acid had no effect on Shannon index of fungal community, but significantly increased Chao1 index. The results indicated that the addition of exogenous phenolic acid could significantly improve the category richness of fungal species, but did not have a good effect on the evenness. Therefore, exogenous phenolic acids did not obvious change the fungal Shannon index (Figure 2B).
Beta Diversity Analysis
The continuous cropping of P. notoginseng significantly [by permutational multivariate analysis of variance (PERMANOVA), P < 0.05] altered the community structure of the soil bacteria and fungi (Figures 3A,B). Exogenous phenolic acids significantly (PERMANOVA, P < 0.05) changed the soil bacterial community structure but had no significant (PERMANOVA, P > 0.05) effect on the fungi (Figures 3C,D).
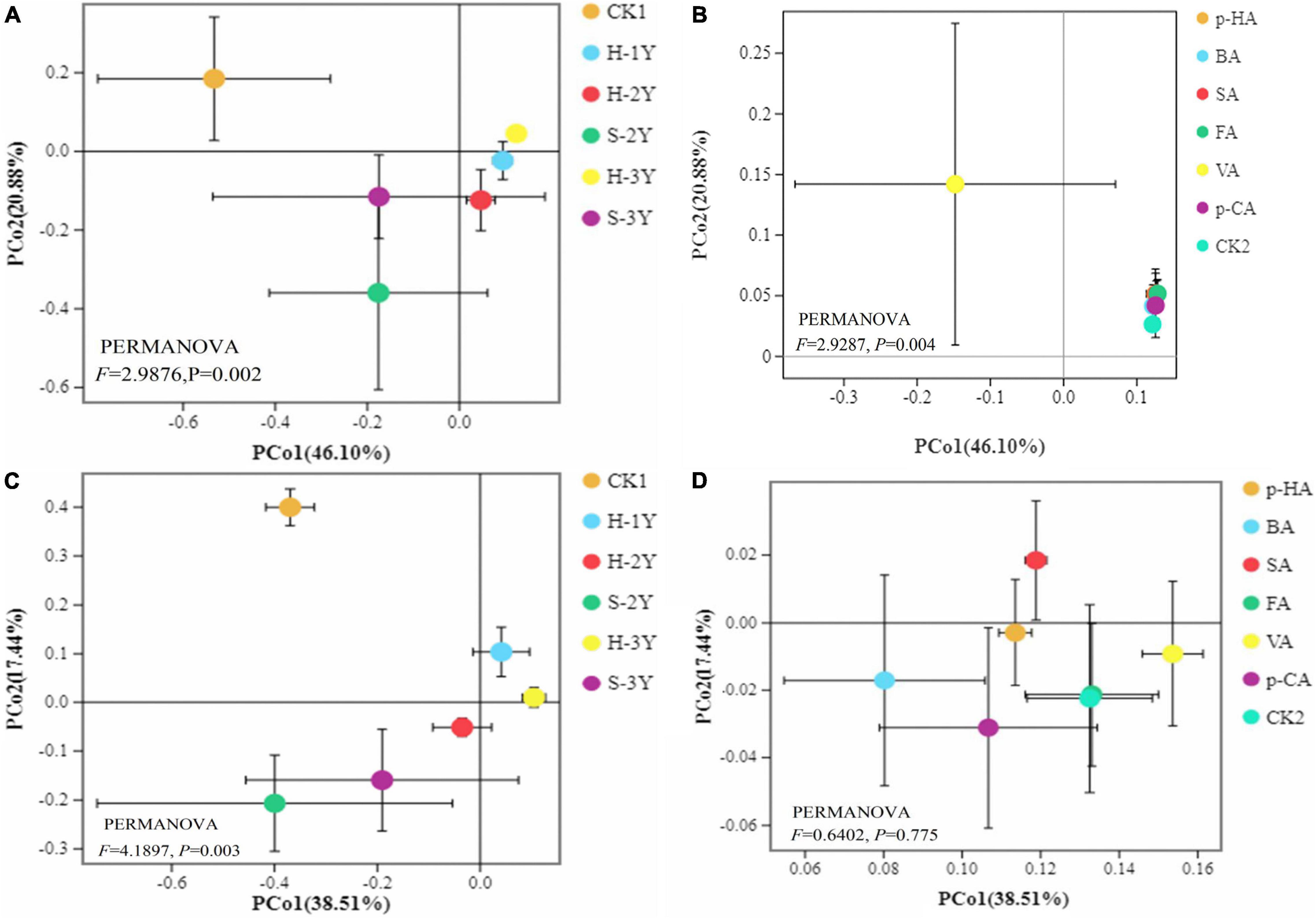
Figure 3. Principal co-ordinate analysis (PCoA). Greenhouse bacterial groups (A), laboratory bacterial group (B), greenhouse fungal groups (C), laboratory fungal group (D). Each point represents a sample. The closer the points are to each other on the plane, the more similar the bacterial community structure of the samples.
Microbiological Specificity Analysis
The specificity of microorganisms in rhizospheric soil decreased after P. notoginseng was planted. The species specificity of diseased P. notoginseng rhizosphere soil was lower than that of healthy soil and control soil (Supplementary Tables 1, 2). Exogenously added phenolic acids changed the species specificity. There were 13 kinds of specific bacteria in VA treatment group, which was much higher than that in control group CK2 (Supplementary Table 1). There were 22 specific fungi in p-HA treatment group, fungal species specificity was increased compared with the control group. The result indicated that p-HA could improve the species specificity of fungi (Supplementary Table 2).
Characteristics of the Rhizospheric Soil Microbial Communities and the Effects of Phenolic Acids
The abundance of microorganisms in the rhizospheric soil was closely related to the number of years of P. notoginseng cultivation. The abundance of Gemmatimonas and Haliangium significant increased, while Lachnospirae-NK4a136-Group and Ruminiclostridium-9 significant decreased in continuously cropped healthy P. notoginseng rhizospheric soil. In rhizospheric sick soil of 2-year-old (S-2Y), the abundance of Lelliottia and Flavobacterium increased significantly (Figure 4A). Flavobacterium is a specific species in the rhizosphere soil of healthy 3-year-old P. notoginseng (Supplementary Table 1). In rhizospheric sick soil of 3-year-old (S-3Y) the abundance of Gemmatimonas increased significantly compared with control soil (Figure 4A). The interaction between allelopathic autotoxins and root microorganisms is the main cause of continuous cropping obstacles (Feng et al., 2020). In terms of bacterial community structure, VA significantly increased the abundance of Ruminiclostridium-9 and Lachnospirae-NK4A136-Group and significantly reduced the abundance of Haliangium. Para-HA, BA, FA, SA significantly increased the abundance of ADurbBin063-1 and Candidatus-Udaeobacter (Figure 4A).
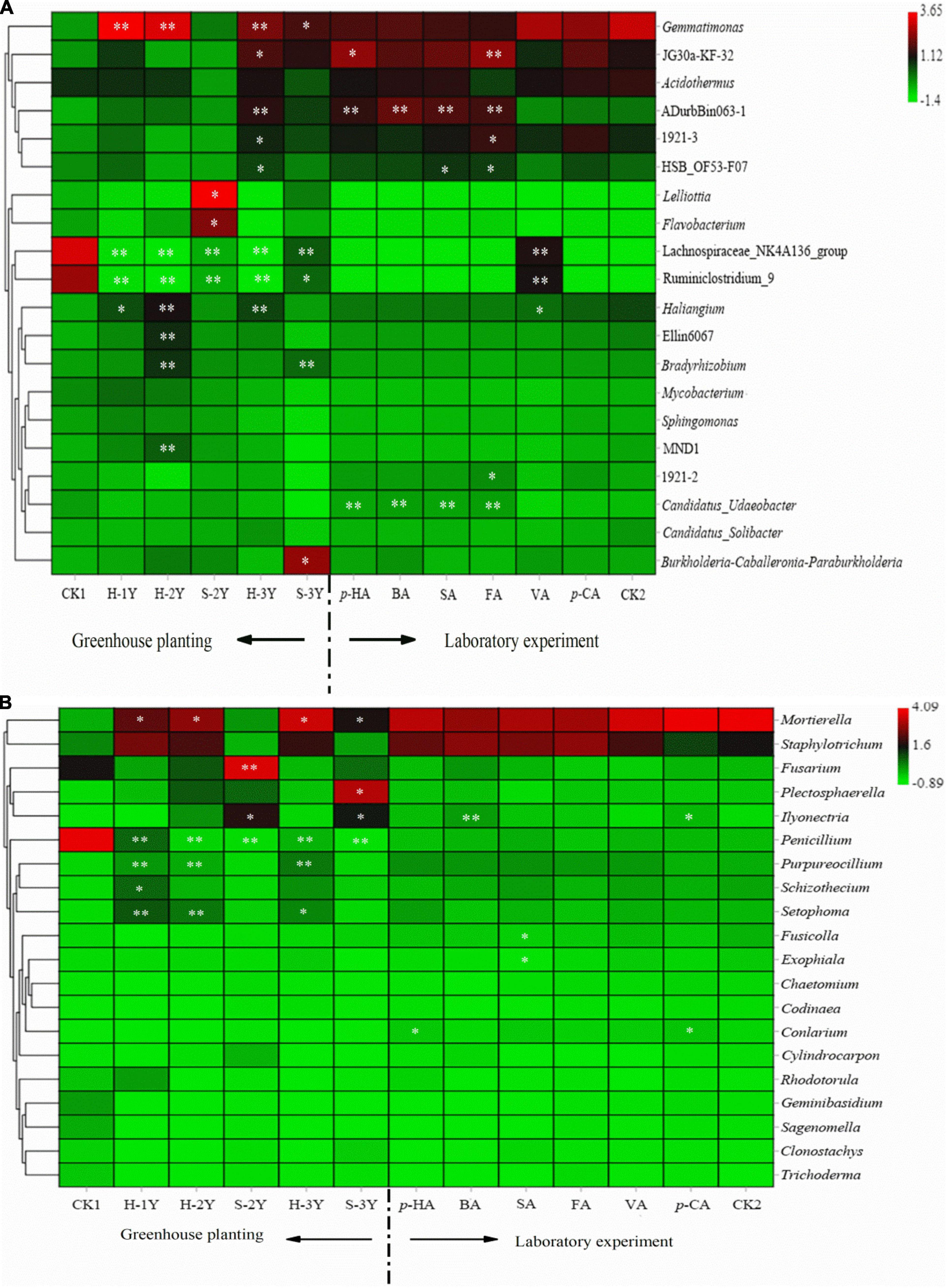
Figure 4. Heat map of bacterial abundances in P. notoginseng rhizospheric soil varying in number of planting years and treated with exogenous phenolic acids (A). Heat map of fungal abundances in P. notoginseng rhizospheric soil varying in number of planting years and treated with exogenous phenolic acids (B). Column normalization was performed for both graphs (A,B). The bacterial sequencing data were compared with SILVA database to obtain bacterial taxa. Fungal sequencing data were compared with UNITE database to obtain fungal taxa. We selected the top 20 species with relative abundance above 0.1% in at least one sample for heat map analysis. CK1 is the natural soil that has not been planted with P. notoginseng, and CK2 is the healthy 3-year-old rhizospheric soil without exogenous phenolic acid addition. The closer the color is to red, the higher the microbial abundance. And the closer the color is to green, the lower the microbial abundance. [* indicates a significant difference from the control group* indicates P < 0.05; ** indicates P < 0.01 (LSD-test)].
Continuous cropping in healthy P. notoginseng caused the abundance of Mortierella, Purpureocillium, and Setophoma increased significantly, while Penicillium decreased significantly. After P. notoginseng was infected, the abundance of Ilyonectria increased significantly. In rhizospheric sick soil of 2-year-old (S-2Y), the abundance of Fusarium increased significantly. In rhizospheric sick soil of 2-year-old (S-3Y), the abundance of Plectosphaerella increased significantly (Figure 5B). Para-HA and VA significant increased the abundance of Ilyonectria. Para-HA and p-CA significantly increased the abundance of Conlarium. SA significantly reduced the abundance of Exophiala and Fusicolla (Figure 4B).
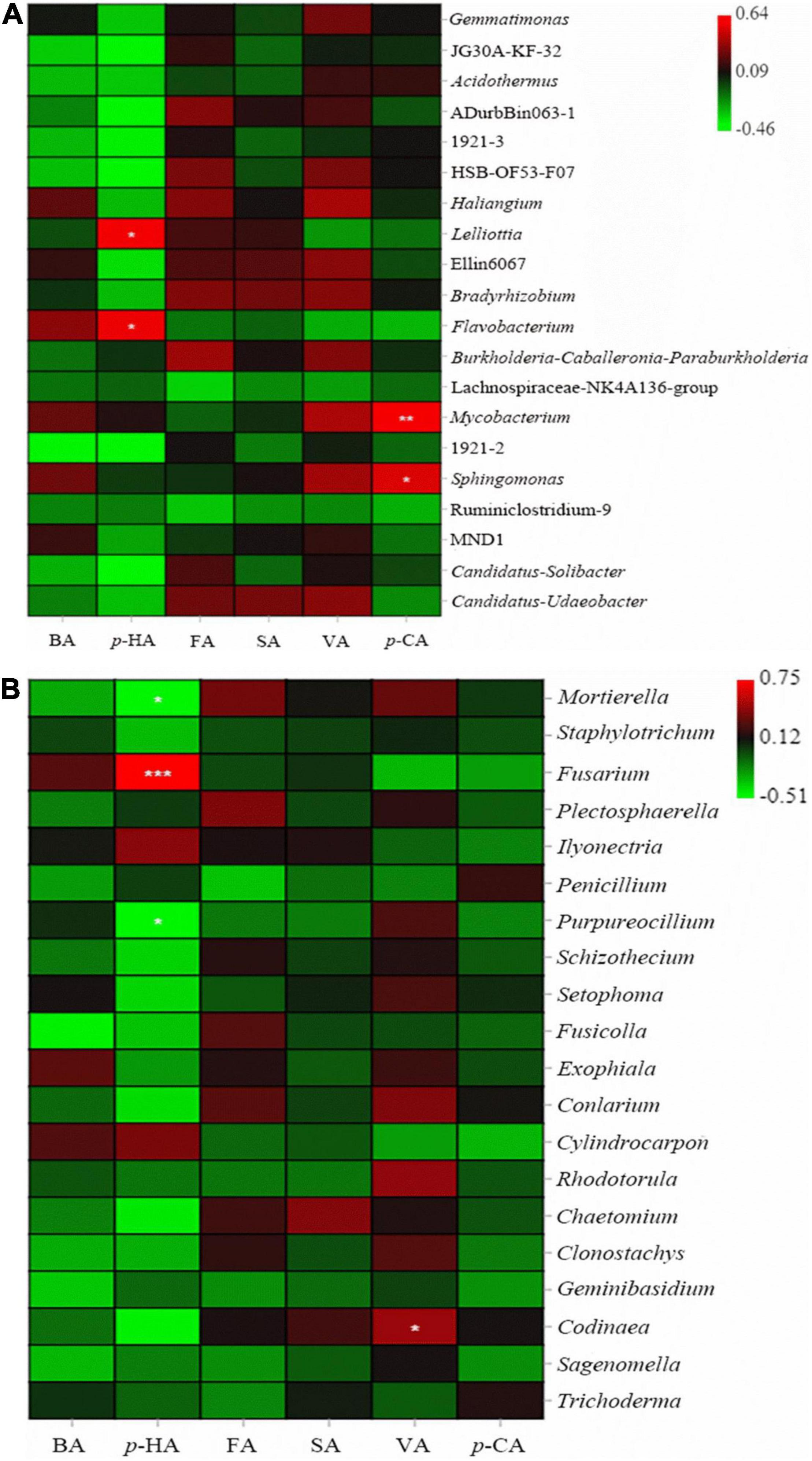
Figure 5. Correlations of phenolic acids with bacteria (A) and fungi (B). The colors in the figure indicate the strength of the correlation. The closer the color is to red, the stronger the positive correlation, while the closer the color is to green, the stronger the negative correlation. * indicates a significant correlation [* indicates P < 0.05; ** indicates P < 0.01; and *** indicates P < 0.001 (Tukey’s test)].
FUNGuild Analysis of Fungi
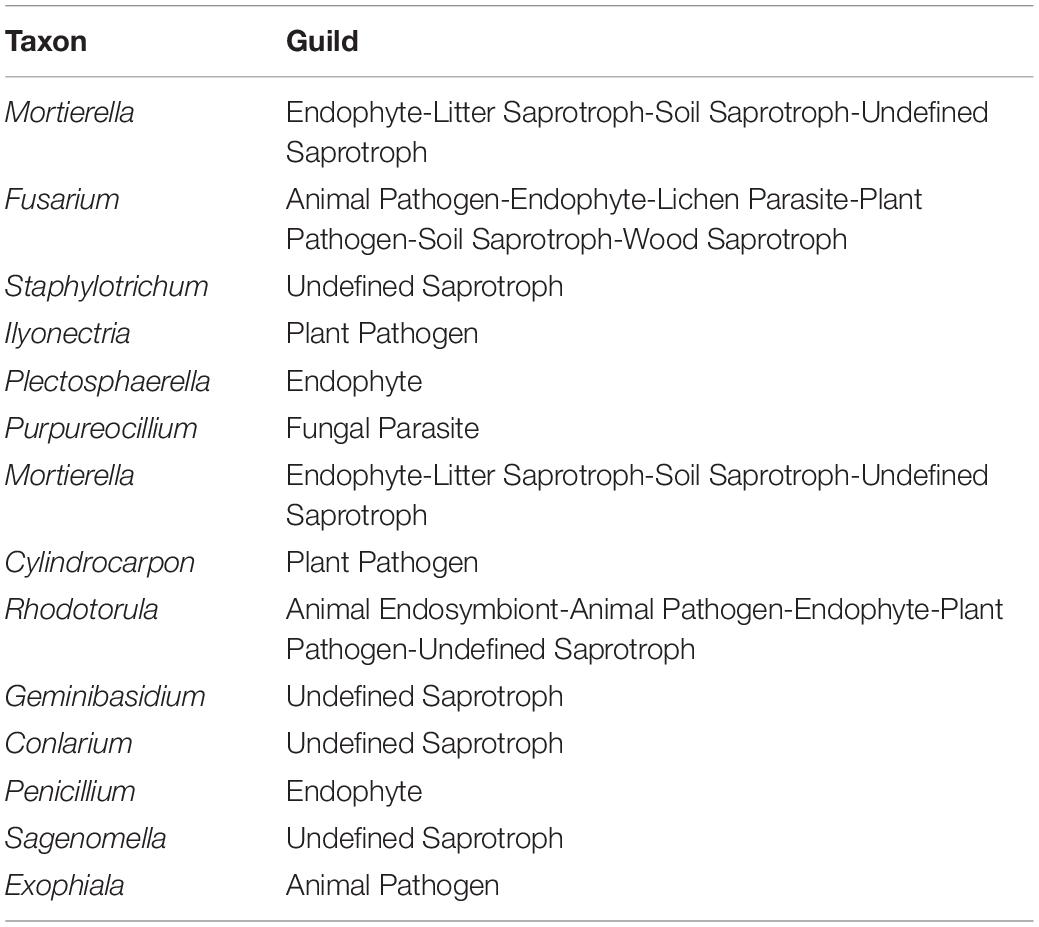
FUNGuild predicted the pathogenicity of 15 species in the top 20 fungi. The genera Fusarium, Ilyonectria, Cylindrocarpon, and Rhodotorula are predicted to be plant pathogens (Table 3).
Analysis of Correlations of Phenolic Acids and Rhizospheric Soil Microorganisms
There was an obvious correlation between phenolic acids and microbial abundance in P. notoginseng rhizospheric soil. An analysis of correlations between phenolic acids and bacteria showed that p-HA was significantly positively correlated with Lelliottia and Flavobacterium. Para-CA was significantly positively correlated with Mycobacterium and Sphingomonas. FA and VA were positively correlated with most bacteria (Figure 5A).
The analysis of correlations between phenolic acids and fungi showed that VA was positively correlated with most fungi, and the other 5 phenolic acids were negatively correlated with most fungi. VA was significantly positively correlated with Codinaea. Para-HA was significantly positively correlated with Fusarium and was significantly negatively correlated with Mortierella and Purpureocillium. Para-HA, BA, FA, and SA were positively correlated with Ilyonectria (Figure 5B).
Degradation of Phenolic Acids by Soil Microorganisms
Six phenolic acids were degraded in total, but the degradation effects of FA and SA were not obvious, so only the degradation curves of p-HA, BA, p-CA, and VA are shown. The degradation of the four phenolic acids by rhizospheric soil microorganisms was clear; degradation was rapid during the first 6 h and tended to slow after 6 h (Figure 6). According to the first-order kinetic equation, the half-lives of p-HA, BA, p-CA, and VA were 2.60, 2.05, 2.28, and 4.41 h, respectively. It follows that the rate of degradation of BA by soil microorganisms was the fastest, and that of VA was the slowest.
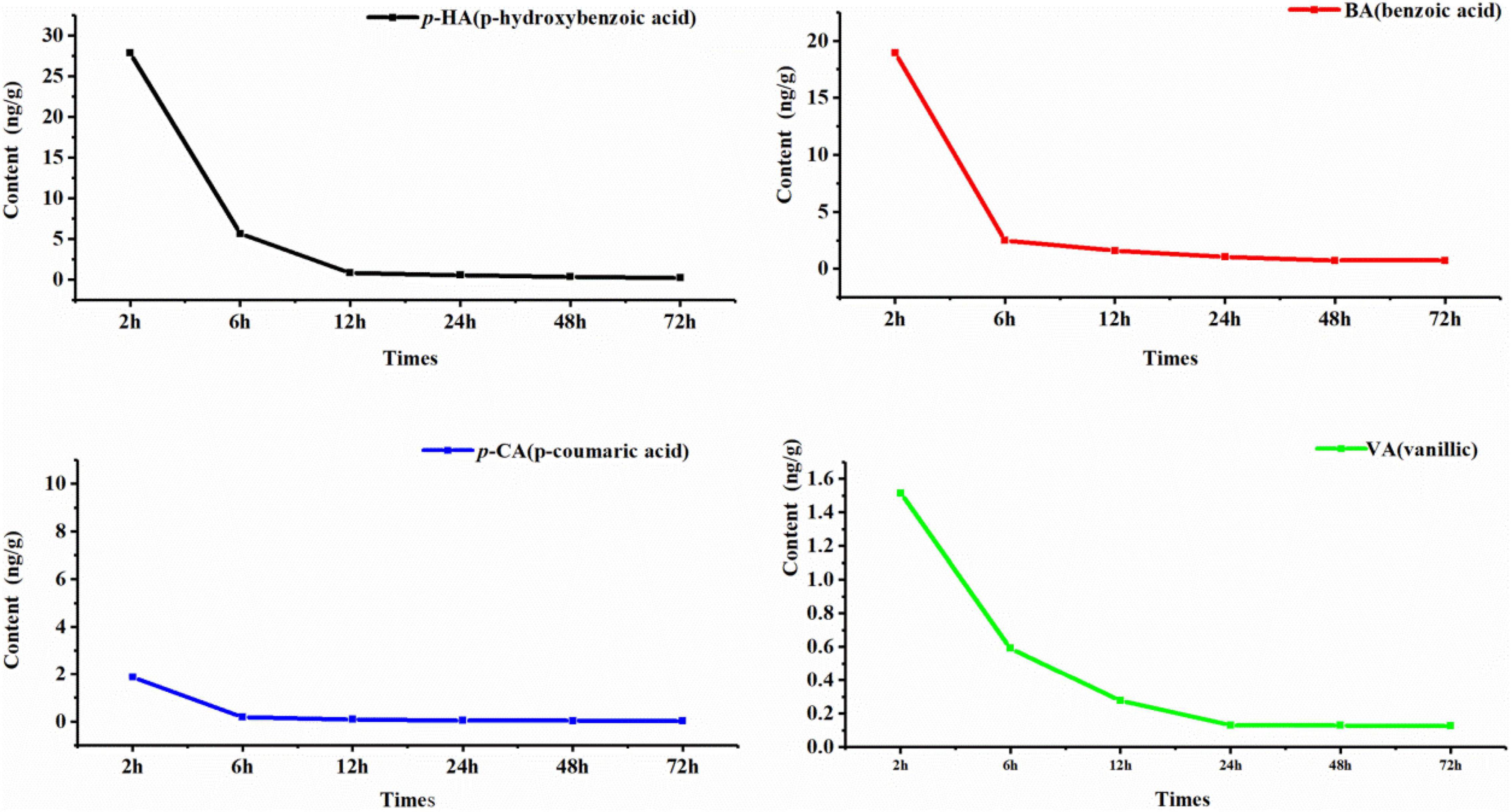
Figure 6. Kinetic degradation curve of phenolic acids. The horizontal column represents the degradation time, and the vertical axis represents the phenolic acid content. The black curve is p-HA, the red curve is BA, the blue curve is p-CA, and the green curve is VA.
Analysis of Correlations of Soil Environmental Factors and Bacterial/Fungal Communities
According to the results of detrended correspondence analysis (DCA), the response of the bacterial community to soil environmental factors was unimodal, while the response of the fungal community was linear. Bacterial communities were examined by canonical correspondence analysis (CCA), and fungal communities were studied by redundancy analysis (RDA). The top 20 dominant species, whose genus names shown in the heat map Figure 4, were selected for the mapping analysis. The blue dots represent these micro flora. AK, TN, HN, and OM exhibited the greatest influence on the bacterial and fungal community (Figures 7A,B). The projection of species on the arrow indicates a positive correlation between abundance of species and environmental factors, while the projection on the backward extension of the arrow indicates a negative correlation. Therefore, TP and AP were positively correlated with the most bacteria. The correlation between soil environmental factors and most fungi was not obvious. In conclusion, AK, TN, HN, and OM exert the greatest influence on rhizospheric soil microorganisms.
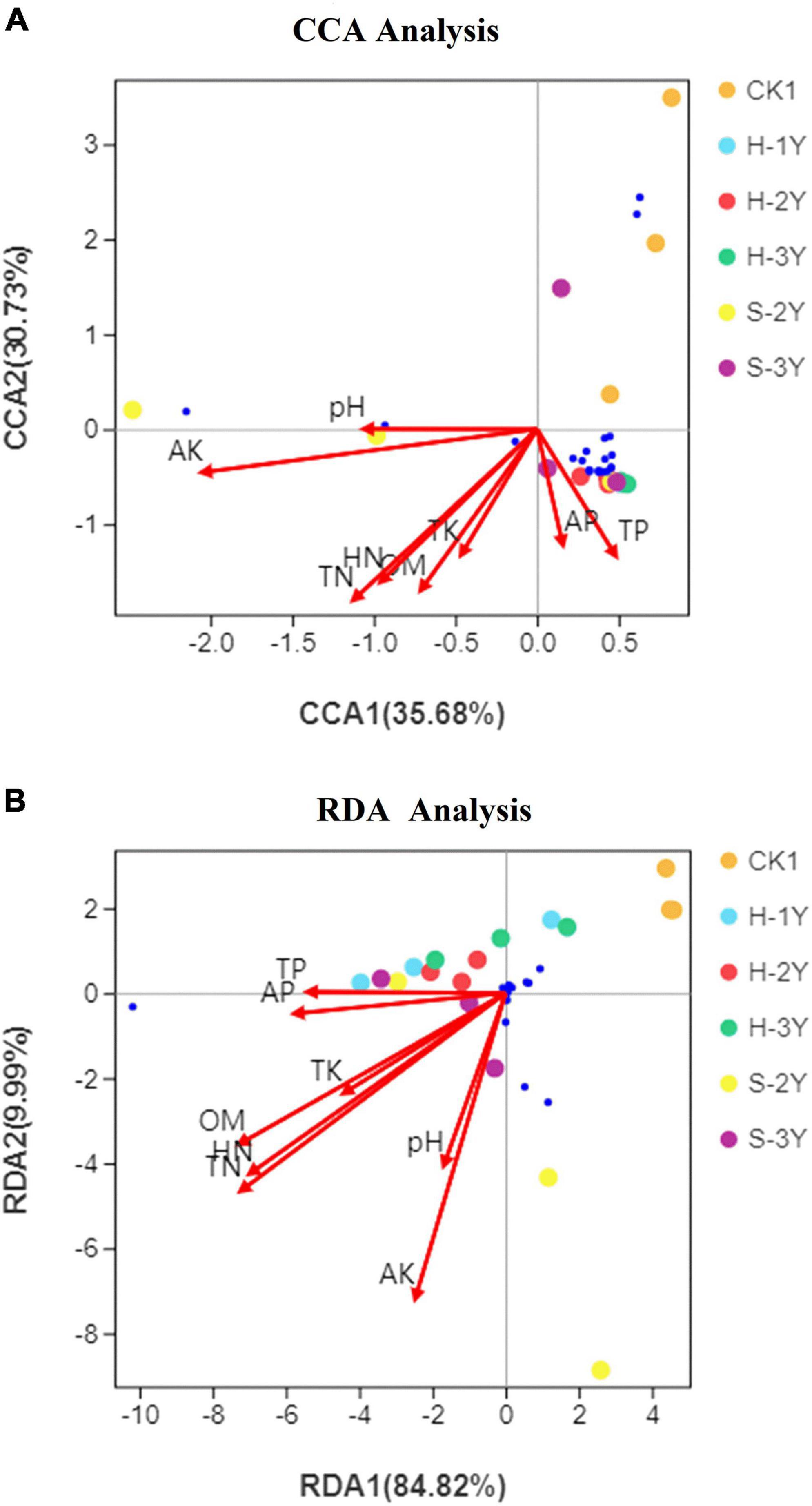
Figure 7. Canonical correspondence analysis of the bacterial community with soil environmental factors (A) and redundancy analysis of soil environmental factors with the fungal community (B). OM is organic matter, TN is total nitrogen, TP is total phosphorus, TK is total potassium, HN is hydrolysable nitrogen, AP is available phosphorus, and AK is available potassium. The top 20 dominant species, whose genus names shown in the heat map Figure 4, were selected for the mapping analysis. The blue dots represent these micro flora.
Discussion
Soil Physical and Chemical Properties Are Related to Microbial Community Structure
The massive use of mineral fertilizers, especially under intensive cultivation systems, appears to exacerbate declines in soil quality by inducing acidification and salinization (Cesarano et al., 2017). Although the degradation of soil quality does not directly cause soil disease in plants, the composition and abundance of soil microorganisms are controlled by soil properties (Paterson et al., 2007). The effects of AK, OM, TN, and HN on the bacterial community were the greatest. Studies have shown that total nitrogen and organic matter had significant effects on bacterial community (Li C. et al., 2020) and the excessive application of nitrogen potash will cause soil salt accumulation and decrease the C/N ratio, resulting in changes to the plant rhizospheric soil bacterial community (He et al., 2017). Also AK, OM, TN, and HN showed the greatest effects on the fungal community. Previous studies showed that the most important factors affecting soil fungal community were organic carbon, available nitrogen, total nitrogen, and C/N ratio (Zhao et al., 2021). This conclusion is similar to the result of this study. Soil microorganisms can sense changes in soil properties caused by nitrogen fertilizer and respond in different ways (Zhao et al., 2014). Nitrogen fertilizer application has been found to reduce the abundances of microorganisms in the rhizospheric soil of plants and reduced the diversity and richness of the community (Shen et al., 2021). In conclusion, soil physical and chemical properties play an important role in the assembly of microbial communities (Tayyab et al., 2019).
Phenolic Acids Associated With Soil Sickness
There are many factors affecting continuous cropping obstacles, such as the accumulation of soil-borne pathogens and parasites, changes in soil microbial community composition, and root exudates (Cesarano et al., 2017). Allelopathic autotoxic substances in the root exudates of P. notoginseng can affect plants by destroying the plant cell structure, affecting plant photosynthesis, and promoting the occurrence of continuous cropping soil sickness (Zhang et al., 2018). In particular, phenolic acids secreted by plant roots can inhibit the germination of seeds and reduce the immunity and stress resistance of plants (Feng et al., 2020). Phenolic acid autotoxins can affect plant growth by inhibiting photosynthesis. The literature indicates that ferulic acid can reduce the chlorophyll content in P. notoginseng seedlings, reduce the photosynthetic rate and inhibit plant growth (Shen et al., 2016). The FA concentration increased with increasing number of P. notoginseng planting years, and reached a maximum value for S-2Y. Para-HA and SA acid contents were highest for S-2Y. Among the collected samples, the S-2Y rhizospheric soil sickness was the most serious. Therefore, FA, p-HA, and SA may be related to soil sickness in P. notoginseng.
Effects of Panax notoginseng Continuous Cropping on Soil Microorganisms
Long-term continuous cropping changes the soil microbial diversity (Sun et al., 2014; Zhao et al., 2014). P. notoginseng continuous cropping increased bacterial Chao1 index and Shannon index and decreased fungal Shannon index. After P. notoginseng disease, bacterial Shannon index reduced and fungal Chao1 index increased. Beta-diversity analysis also showed that continuous cropping significantly changed the microbial community structure. In addition, the specificity of the microbial community was also changed by continuous cropping and was decreased after disease. Abundances of some microbial taxa associated with P. notoginseng also changed after continuous cropping. The results showed that continuous P. notoginseng cropping changed the soil microbial community and resulted in continuous cropping soil sickness. A large number of studies have shown that continuous cropping leads to the accumulation of organic and phenolic acids and affects the microbial community structure in rhizospheric soil (Blagodatskaya and Kuzyakov, 2013).
Effects of Phenolic Acids on Soil Microorganisms
The number and species of rhizospheric soil microorganisms are important factors affecting plant growth, development and health (Berendsen et al., 2012; Yang et al., 2017). Phenolic acids exert direct effects on crops but can also indirectly affect microbial biomass, community structure and biodiversity (Qu and Wang, 2008; Zhou and Wu, 2012). Phenolic acids reduce the uniformity of fungi, which is beneficial to the enrichment of some fungi, but this is not beneficial for plants. Because most plant diseases are caused by fungi, the accumulation of rhizospheric fungi may exacerbate continuous cropping soil sickness (Zhang et al., 2020). Beta-diversity analysis showed that phenolic acids could significantly change bacterial community structure. VA in significantly reduced the bacterial Shannon index. Exogenous p-HA, FA, SA, and VA significantly increased the fungal Chao1 index and p-HA showed the most significant effect. There is growing evidence that plants can alter the soil microbiota by secreting bioactive molecules into the rhizosphere and that the microbiome can also be modified by root exudates to form host-specific microbial communities (Luo et al., 2020). VA affected bacterial specificity, and p-HA affected fungal specificity. Thus, VA and p-HA affect both microbial diversity and microbial specificity, and it is a matter of time until a specific community that is dependent on these two phenolic acids will form. Phenolic acids can change the soil microbial community structure and cause negative feedback between plants and microorganisms, which can cause toxic effects on host plants and aggravate continuous cropping obstacles. Phenolic acids can change soil microbial community structure and cause soil microbial imbalance, resulting in toxic effects on host plants, thus inducing continuous cropping soil sickness (Zhou et al., 2018; Jin et al., 2020).
The phenolic acids in P. notoginseng rhizospheric soil are clearly correlated with microbial community distribution (Yang et al., 2020). VA is positively correlated with most fungi and bacteria, indicating that VA can promote the growth of rhizospheric microbes (Wu et al., 2016). Para-HA was positively correlated with an increase in bacteria (Lelliottia and Flavobacterium) after P. notoginseng disease. This result indicates that p-HA was able to promote the growth of these two bacteria. The main sign of continuous cropping soil sickness in P. notoginseng is root rot, which is caused by fungi such as Fusarium and Cylindrocarpon destructans (Guo et al., 2009; Wen et al., 2020). FUNGuild also predicted that Ilyonectria was a plant pathogen. Para-HA was positively correlated with Fusarium and Ilyonectria. In conclusion, p-HA and VA are the main autotoxic phenolic acids in P. notoginseng continuous cropping.
Soil Microorganisms Degrade Phenolic Acids
Previous studies have found that the rhizospheric environment and root exudates of plants interact with each other. On the one hand, changes in the rhizosphere affect the types and quantities of exudates; on the other hand, rhizosphere exudates also cause changes in the rhizosphere (Zhang et al., 2007). The half-life periods of p-HA, BA and p-CA were all less than 3 h, while the half-life of VA was less than 4.5 h. Thus, soil microorganisms can degrade phenolic acids rapidly. It is inferred that if the microbial community of P. notoginseng roots changes, the phenolic acid content of the P. notoginseng rhizospheric soil will increase or decrease accordingly.
Suggestions for Solving Panax notoginseng Continuous Cropping Soil Sickness
Continuous cropping soil sickness in P. notoginseng urgently needs to be solved. Rhizospheric microecology involves a special interaction between crop-soil microorganisms and the environment. Crop root exudates and soil microorganisms interact in the soil, which jointly affects the growth and development of crops. Coordinating the relationships among these factors is the key to solving the problems associated with continuous cropping (Fu and Jia, 2012). This study creatively combined field investigation with laboratory experiments to determine the main phenolic acid autotoxins (p-HA and VA). Exogenously supplied phenolic acids were able to affect the diversity and abundance of soil microbes. There were strong correlations between phenolic acids and rhizospheric soil microorganisms. Soil microorganisms can also degrade phenolic acids quickly. It is a dynamic equilibrium. Therefore, maintaining a balance between phenolic acids and soil microorganisms can alleviate continuous cropping obstacles. Crop rotation and fallowing can also be used to balance the interaction between soil autotoxic substances and microorganisms before cultivation. Fallowing and crop rotation could maintain the dynamic balance between phenolic acids and soil microorganisms. Soil diseases can also be alleviated by adding beneficial microorganisms that can degrade phenolic acids to the soil.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA757903.
Author Contributions
LB was responsible for experimental operation and manuscript writing. YL and JS were responsible for the collection of experimental samples. YD was responsible for solving technical problems. YT was responsible for the design of the experimental method and solving the problems encountered during the experiment. YW and FZ played a supervisory role. All authors contributed to the article and approved the submitted version.
Funding
This work was supported financially by the National Natural Science Foundation of China (Nos. 31960630 and 31960232).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to Guangzhou Gene Denovo Biotechnology Co., Ltd. for assisting with the sequencing and bioinformatics analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.791603/full#supplementary-material
References
Berendsen, R. L., Pieterse, C., and Bakker, P. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Blagodatskaya, E., and Kuzyakov, Y. (2013). Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol. Biochem. 67, 192–211. doi: 10.1016/j.soilbio.2013.08.024
Bremner, J. M. (1960). Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 55, 11–33. doi: 10.1017/S0021859600021572
Cesarano, G., Zotti, M., Antignani, V., Marra, R., Scala, F., and Bonanomi, G. (2017). Soil sickness and negative plant-soil feedback: a reappraisal of hypotheses. J. Plant Pathol. 99, 545–570. doi: 10.4454/jpp.v99i3.3960
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Commission, S. P. (2015). Pharmacopoeia of the People’s Republic of China:Part I. Beijing: China Medical Science and Technology press, 11–12.
Dong, L., Xu, J., Feng, G., Li, X., and Chen, S. (2016). Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci. Rep. 6:31802. doi: 10.1038/srep31802
Dong, L., Xu, J., Li, Y., Fang, H., Niu, W., Li, X., et al. (2018). Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol. Biochem. 125, 64–74. doi: 10.1016/j.soilbio.2018.06.028
Feng, J., Zhou, B., Liu, Q., Wang, X., Lu, H., Guo, L., et al. (2020). Study on the relationship between rhizosphere microorganisms and phenolic acids and its countermeasures. China J. Trad. Chin. Med. Pharm. 35, 4049–4052.
Fu, J., and Jia, Z. (2012). Research progress on the mechanism and regulation of continuous cropping obstacle of flue-cured tobacco. Anhui Agric. Sci. 40:13762. doi: 10.3969/j.issn.0517-6611.2012.28.030
Gu, M., Xu, W., Mao, J., Zhang, Z., Tang, G., and Ge, C. (2012). Microbial community diversity of rhizosphere soil in continuous cotton cropping system in Xinjiang. Acta Ecol. Sin. 32, 3031–3040. doi: 10.5846/stxb201104170503
Guo, R., Liu, X., Li, S., and Miao, Z. (2009). In vitro inhibition of fungal root-rot pathogens of Panax notoginseng by rhizobacteria. Plant Pathol. J. 25, 70–76. doi: 10.5423/PPJ.2009.25.1.070
Guo, Z.-Y., Kong, C.-H., Wang, J.-G., and Wang, Y.-F. (2011). Rhizosphere isoflavones (daidzein and genistein) levels and their relation to the microbial community structure of mono-cropped soybean soil in field and controlled conditions. Soil Biol. Biochem. 43, 2257–2264. doi: 10.1016/j.soilbio.2011.07.022
Gyaneshwar, P., Kumar, G. N., Parekh, L. J., and Poole, P. S. (2002). Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245, 83–93. doi: 10.1023/a:1020663916259
He, Z., Lou, C., Wang, X., Dong, H., and Han, Y. (2017). Nitrogen and potassium on soil microbial community of tomato and soil nutrient and salinity. Northern Hortic. 9, 148–153. doi: 10.11937/bfyy.201709032
Jia, M., Li, Q., Hua, J., Liu, J., Zhou, W., Qu, B., et al. (2020). Phytohormones regulate both “Fish Scale” galls and cones on Picea koraiensis. Front. Plant Sci. 11:580155. doi: 10.3389/fpls.2020.580155
Jin, X., Wu, F., and Zhou, X. (2020). Different toxic effects of ferulic and p-hydroxybenzoic acids on cucumber seedling growth were related to their different influences on rhizosphere microbial composition. Biol. Fertil. Soils 56, 125–136. doi: 10.1007/s00374-019-01408-0
Kuczynski, J., Stombaugh, J., Walters, W. A., Gonzalez, A., Caporaso, J. G., and Knight, R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. Chapter 1, Unit1E.5.–Unit1E.5. doi: 10.1002/9780471729259.mc01e05s27
Li, C., Liu, Y., and Liu, J. (2020). Analysis factors of rhizosphere bacterial community diversity in three genuine medical plants. J. Ecol. Environ. 10, 1988–1993. doi: 10.16258/j.cnki.1674-5906.2020.10.008
Li, X., Ding, C., Hua, K., Zhang, T., Zhang, Y., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi: 10.1016/j.soilbio.2014.07.019
Li, Y., Liu, X., Wu, D., Chen, B., Ren, X., and Tang, J. (2020). Effects of continuous cropping of greenhouse cucumber on soil fungal abundance and community structure Province. Acta Agric. Boreali Sin. 35, 194–204. doi: 10.7668/hbnxb.20190712
Li, Y., Ying, Y., Zhao, D., and Ding, W. (2014). Influence of allelochemicals on microbial community in ginseng cultivating soil. Chin. Herb. Med. 6, 313–318. doi: 10.1016/s1674-6384(14)60047-2
Li, Z., Fu, J., Zhou, R., and Wang, D. (2018). Effects of phenolic acids from ginseng rhizosphere on soil fungi structure, richness and diversity in consecutive monoculturing of ginseng. Saudi J. Biol. Sci. 25, 1788–1794. doi: 10.1016/j.sjbs.2018.07.007
Liu, P., Zhao, H., Li, Q., Lin, H., Shen, Y., and Wan, S. (2018). Effects of three phenolic acid allelochemicals on rhizosphere soil microbes and pod yield of peanut (Arachis hypogaea L.). Chin. J. Oil Crop Sci. 40, 101–109. doi: 10.7505/j.issn.1007-9084.2018.01.013
Liu, X., and Li, X. (2021). Basic biostatistical tests and their R codes. Chin. J. Appl. Entomol. 58, 220–232. doi: 10.7679/j.issn.2095-1353.2021.024
Luo, L., Yang, L., Yan, Z., Jiang, B., Li, S., Huang, H., et al. (2020). Ginsenosides in root exudates of Panax notoginseng drive the change of soil microbiota through carbon source different utilization. Plant Soil 455, 139–153. doi: 10.1007/s11104-020-04663-5
Macdermot, H. E. (1950). Pharmaceutical botany. Can. Med. Assoc. J. 147:613. doi: 10.1001/jama.1951.03670230079034
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mclean, E. O., and Watson, M. E. (1985). “Soil measurements of plant-available potassium,” in Potassium in Agriculture, ed. R. D. Munson (Madison, WI: ASA, CSSA, and SSSA), 277–308. doi: 10.2134/1985.potassium.c10
Neefs, J. M., Peer, Y., Rijk, P. D., Goris, A., and Wachter, R. D. (1991). Compilation of small ribosomal subunit RNA sequences. Nucl. Acids Res. 19 Suppl (Suppl), 1987–2015. doi: 10.1093/nar/19.suppl.1987
Nelson, D. W., and Sommers, L. E. (1996). Total carbon, organic carbon, and organic matter. Methods Soil Anal. 34, 539–579. doi: 10.2136/sssabookser5.3.c34
Paterson, E., Gebbing, T., Abel, C., Sim, A., and Telfer, G. (2007). Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610. doi: 10.1111/j.1469-8137.2006.01931.x
Qian, P., Mi, C., Jiang, K., Yang, R., Li, O., and Hu, X. (2016). Phylogenetic and metabolic responses of rhizosphere microbes to the cultivation of Panax notoginseng. J. Biobased Mater. Bioenerg. 10, 370–377. doi: 10.1166/jbmb.2016.1616
Qiao, Y., Gu, C., Zhu, H., Wang, D., Zhang, M., Zhang, Y., et al. (2020). Allelochemicals of Panax notoginseng and their effects on various plants and rhizosphere microorganisms. Plant Divers. 42, 323–333. doi: 10.1016/j.pld.2020.04.003
Qiu, M., Zhang, R., Xue, C., Zhang, S., Li, S., Zhang, N., et al. (2012). Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 48, 807–816. doi: 10.1007/s00374-012-0675-4
Qu, X. H., and Wang, J. G. (2008). Effect of amendments with different phenolic acids on soil microbial biomass, activity, and community diversity. Applied Soil Ecol. 39, 172–179. doi: 10.1016/j.apsoil.2007.12.007
Riley, D., and Barber, S. A. (1970). Salt accumulation at the soybean (glycine max. (L.) Merr.) root-soil interface1. Soil Sci. Soc. Am. J. 34, 154–155. doi: 10.2136/sssaj1970.03615995003400010042x
Shen, J., Zhang, M., Liu, G., Li, X., Shi, D., and Wang, Y. (2021). Effects of lime nitrogen application on microbial flora of tobacco rhizosphere soil. Soil Fertil. Sci. 1, 75–82. doi: 10.11838/sfsc.1673-6257.19562
Shen, Y., Zhang, H., Zhang, Z., and Gao, M. (2016). Allelopathic effects of phenolic acids on Panax notoginseng seedlings. Guangxi Plants 36, 607–614. doi: 10.11931/guihaia.gxzw201507008
Shen, Z., Zhang, Q., Liu, L., and Qiu, Y. (2011). Determination of available phosphorus in soil by sodium bicarbonate extraction Mo-Sb anti-spectrophotometry method. Environ. Monit. Forewarn. 3, 12–15. doi: 10.3969/j.issn.1674-6732.2011.05.004
Sun, J., Zhang, Q., Zhou, J., and Wei, Q. (2014). Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS One 9:e111744. doi: 10.1371/journal.pone.0111744
Tan, Y., Cui, Y., Li, H., Kuang, A., Li, X., Wei, Y., et al. (2017). Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 194, 10–19. doi: 10.1016/j.micres.2016.09.009
Tang, B., Dong, Y., He, M., Liu, J., Wu, K., Guan, H., et al. (2020). Effects of different planting years of healthy Panax notoginseng on the rhizosphere microbial community in Wenshan of Yunnan province. J. Microbiol. 9, 2857–2866. doi: 10.13344/j.microbiol.china.200192
Tayyab, M., Islam, W., Lee, C. G., Pang, Z., Khalil, F., Lin, S., et al. (2019). Short-term effects of different organic amendments on soil fungal composition. Sustainability 11, 1–13. doi: 10.3390/su11010198
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863
Wen, Y., Hunag, H., Ma, Z., Zeng, D., Fu, J., and Zeng, C. (2020). Illumina highthroughput sequencing reveals fungal community composition and diversity in root and rhizosphere soil of healthy and root-rot Panax notoginseng. Food Ferment. Sci. Technol. 56, 22–30. doi: 10.3969/j.issn.1674-506X.2020.06-004
Wu, H., and Lin, W. (2020). A commentary and development perspective on the consecutive monoculture problems of medicinal plants. Chin. J. Eco Agric. 28, 775–793. doi: 10.13930/j.cnki.cjea.190760
Wu, H., Wu, L., Zhu, Q., Wang, J., Qin, X., Xu, J., et al. (2017). The role of organic acids on microbial deterioration in the Radix pseudostellariae rhizosphere under continuous monoculture regimes. Sci. Rep. 7:3497. doi: 10.1038/s41598-017-03793-8
Wu, L., Liu, J., Wang, W., Dai, D., Cheng, X., Zhang, Z., et al. (2014). Identification and content determination of phenolic acids of rhizosphere soil of Panax notoginseng. World Sci. Technol. Modern. Trad. Chin. Med. Mater. Med. 16, 825–829. doi: 10.11842/wst.2014.04.026
Wu, L., Wang, J., Huang, W., Wu, H., Chen, J., Yang, Y., et al. (2016). Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 5:15871. doi: 10.1038/srep15871
Xia, Z., Kong, C., Chen, L., and Wang, S. (2015). Allelochemical-mediated soil microbial community in long-term monospecific Chinese fir forest plantations. Appl. Soil Ecol. 96, 52–59. doi: 10.1016/j.apsoil.2015.07.012
Xing, H., Xiao, Z., Yan, J., Ma, J., and Meng, Y. (2011). Effects of continuous cropping of maize on soil microbes and main soil nutrients. Pratacult. Sci. 28, 1777–1780.
Xu, Y., Tan, H., Li, S., Wang, N., and Feng, Y. (2018). Panax notoginseng for inflammation-related Chronic diseases: a review on the modulations of multiple pathways. Am. J. Chin. Med. 46, 971–996. doi: 10.1142/S0192415X18500519
Yang, H., Li, J., Xiao, Y., Gu, Y., Liu, H., Liang, Y., et al. (2017). An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 8:2179. doi: 10.3389/fmicb.2017.02179
Yang, J., Guan, H., Liu, D., Sun, Y., Wei, M., and Wang, Y. (2016). Study on mechanism of Sanqi (Panax notoginseng F.H.Chen) replant failure and its alleviation technology. North. Hortic. 14, 160–167. doi: 10.11937/bfyy.201614040
Yang, J., Sun, Y., Wei, M., Wang, Y., Huang, T., Liu, Y., et al. (2012). The effect on growth of Panax notoginseng in soil under rotation for the different years. Res. Pract. Chin. Med. 26, 6–8. doi: 10.13728/j.1673-6427.2012.02.005
Yang, M., He, M., Duan, J., Zheng, Y., Wang, J., Zhong, Y., et al. (2020). Effects of biochar addition on phenolic acids and microbial community in rhizosphere soil at continuous cropping field of tobacco. Fujian J. Agric. Sci. 35, 103–110. doi: 10.19303/j.issn.1008-0384.2020.01.014
Zhang, J., Fan, S., Qin, J., Dai, J., Zhao, F., Gao, L., et al. (2020). Changes in the microbiome in the soil of an american ginseng continuous plantation. Front. Plant Sci. 11:572199. doi: 10.3389/fpls.2020.572199
Zhang, X., Li, T., and Wang, Y. (2007). Relationship between plant growth environment and root exudates. Chin. J. Soil Sci. 38, 785–789. doi: 10.1016/S1872-2075(07)60055-7
Zhang, Y., Chen, Y., Lei, F., Li, S., Shi, F., Dou, M., et al. (2018). Advances in research on allelopathic autotoxicity effects of medicinal plants. Chin. Trad. Herb. Drugs 49, 1946–1956. doi: 10.7501/j.issn.0253-2670.2018.08.032
Zhang, Y., Zheng, Y., Xia, P., Xun, L., and Liang, Z. (2019). Impact of continuous Panax notoginseng plantation on soil microbial and biochemical properties. Sci. Rep. 9:13205. doi: 10.1038/s41598-019-49625-9
Zhao, C., Wang, W., Zhang, Y., Xue, X., Luo, X., Luo, L., et al. (2021). Characteristics of soil fungal community structure among different parent materials in rubber plantation. J. South. Agric. 52, 1869–1876. doi: 10.3969/j.issn.2095-1191.2021.07.016
Zhao, J., Ni, T., Li, Y., Xiong, W., Ran, W., Shen, B., et al. (2014). Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS One 9:e85301. doi: 10.1371/journal.pone.0085301
Zhong, Y., Cao, J., Zheng, Y., Wang, J., Xu, Y., Tong, W., et al. (2020). Regulation of microbial agent on soil phenolic acid contents and microbial population. Southwest China J. Agric. Sci. 33, 2037–2041. doi: 10.16213/j.cnki.scjas.2020.9.023
Zhou, X., and Wu, F. (2012). p-Coumaric Acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f.sp. cucumerinum Owen. PLoS One 7:e48288. doi: 10.1371/journal.pone.0048288
Zhou, X., Yu, G., and Wu, F. (2012). Responses of soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.) to exogenously applied p-Hydroxybenzoic acid. J. Chem. Ecol. 38, 975–983. doi: 10.1007/s10886-012-0156-0
Keywords: P. notoginseng, continuous cropping soil sickness, microbial diversity, phenolic acids, autotoxicity
Citation: Bao L, Liu Y, Ding Y, Shang J, Wei Y, Tan Y and Zi F (2022) Interactions Between Phenolic Acids and Microorganisms in Rhizospheric Soil From Continuous Cropping of Panax notoginseng. Front. Microbiol. 13:791603. doi: 10.3389/fmicb.2022.791603
Received: 08 October 2021; Accepted: 03 February 2022;
Published: 24 February 2022.
Edited by:
Wei Zhang, Michigan State University, United StatesReviewed by:
Xingang Zhou, Northeast Agricultural University, ChinaSarah Lebeis, Michigan State University, United States
Copyright © 2022 Bao, Liu, Ding, Shang, Wei, Tan and Zi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Tan, tanyong551@163.com
 Limei Bao
Limei Bao Yuyan Liu
Yuyan Liu Yafang Ding2
Yafang Ding2 Yong Tan
Yong Tan