- 1Yunnan Herbal Laboratory, School of Ecology and Environmental Science, Yunnan University, Kunming, China
- 2The International Joint Research Center for Sustainable Utilization of Cordyceps Bioresources in China and Southeast Asia, Yunnan University, Kunming, China
- 3Faculty of Agricultural Technology, Rajamangala University of Technology, Thanyaburi, Thailand
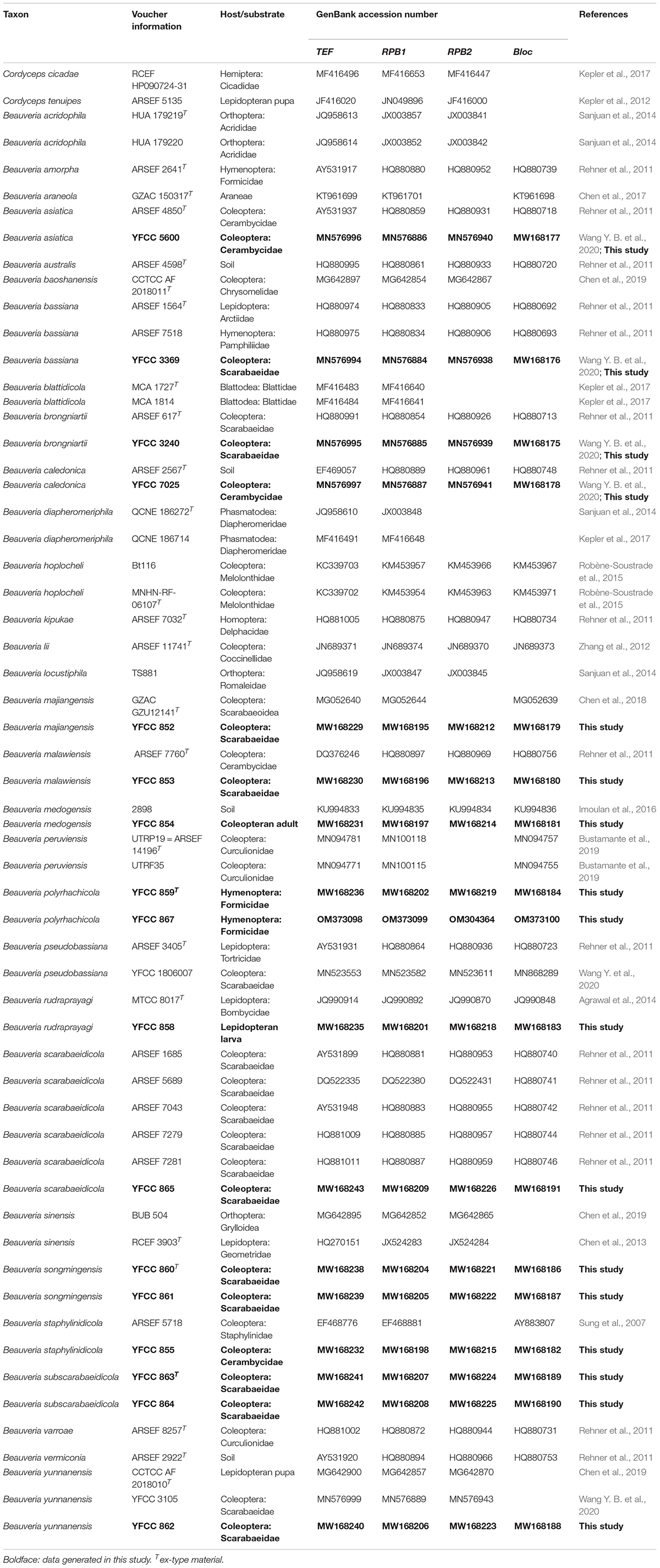
Beauveria is a very important fungal resource. Some Beauveria species have great economic and ecological value. Through surveying Beauveria in China and Thailand over the past 4 years, 15 Beauveria spp. were collected and identified. Three new species—namely, B. polyrhachicola, B. songmingensis, and B. subscarabaeidicola—were described and illustrated based on morphological characteristics and molecular data. The phylogenetic positions of the 15 species were evaluated according to phylogenetic inferences based on six loci (nrSSU, nrLSU, TEF, RPB1, RPB2, and Bloc). Nine species of Beauveria in our study were isolated from adult scarab beetles (Coleoptera: Scarabaeidae). The pathogenicity of the isolates from the B. bassiana complex and B. scarabaeidicola complex was determined with three bioassays using B. mori and T. molitor larvae, in addition to Protaetia brevitarsis adults. The results indicated that the B. bassiana complex isolates had great potential in the biocontrol of the three insects; by contrast, the B. scarabaeidicola complex isolates showed obvious host specificity and low virulence.
Introduction
Beauveria is a very important fungal resource, with some species having great economic and ecological value (Zimmermann, 2007; Rehner et al., 2011; Wang Y. et al., 2020). Beauveria bassiana (Bals.-Criv.) Vuill. and B. brongniartii (Sacc.) Petch are well-known environmentally safe alternatives to using chemical pesticides to control agricultural pests (Zimmermann, 2007; Rehner et al., 2011). Beauveria pseudobassiana S.A. Rehner and Humber has also been shown to have great potential in the biocontrol of numerous insect pests (Wang Y. et al., 2020). The entomopathogenic fungi Beauveria spp. are a class of environmentally friendly fungal pathogens that play an important role in controlling insect populations in nature (Luo et al., 2018; McKinnon et al., 2018). Some Beauveria species, as endophytes or soil and rhizosphere inhabitants, have been considered for potential use as biocontrol agents against plant pathogens by concerned practitioners, such as agriculturalists and plant pathologists. These species can produce an array of bioactive metabolites that limit the growth of some fungal plant pathogens and induce plant systemic resistance against the pathogenic bacterium (Ownley et al., 2010).
Beauveria bassiana is the most widely used fungus available commercially for controlling agricultural and forestry pests (Li et al., 2011). Products based on this species have been developed in many countries around the world (Goettel et al., 2005; Faria and Wraight, 2007; Li et al., 2011). However, a growing body of molecular evidence has demonstrated that B. bassiana, originally known as a generalist with a global distribution, encompasses cryptic lineages adapted to specific hosts or ecologies (Li et al., 2011; Rehner et al., 2011). Many initially identified B. bassiana isolates may belong to any of the species in the B. bassiana complex, such as B. rudraprayagi Y. Agrawal, Mual and Shenoy, B. staphylinidicola (Kobayasi and Shimizu) B. Shrestha, Kepler and Spatafora, and B. peruviensis D.E. Bustamante, M.S. Calderon, M. Oliva, and S. Leiva (Rehner et al., 2011; Agrawal et al., 2014; Kepler et al., 2017; Bustamante et al., 2019). Therefore, the abovementioned mycoinsecticide formulations of B. bassiana are not likely all based on B. bassiana.
Beauveria scarabaeidicola (Kobayasi) S.A. Rehner and Kepler is widely distributed in Oceania and Asia and named after its host adult beetle (Coleoptera: Scarabaeidae). It was originally described as Cordyceps scarabaeicola occurring in its sexual morph on an adult scarab beetle in New Guinea (Kobayasi and Shimizu, 1976). Cordyceps scarabaeicola has also been reported occasionally from many Asian countries, including China, Japan, and Korea (Shrestha et al., 2014). In an important phylogenetic study of Beauveria, a new entomopathogenic species, B. sungii S.A. Rehner and R.A. Humber, was described as a scarab-killing pathogen (hosts of all B. sungii isolates were identified as scarabs) (Rehner et al., 2011). Later, however, Shrestha et al. (2014) demonstrated that the telemorphic stage of B. sungii was C. scarabaeicola based on morphological and phylogenetic evidence. Because C. scarabaeicola was described earlier than B. sungii, Kepler et al. (2017) recommended B. scarabaeicola as the name of this species. Recently, Chen et al. (2019) proposed a new species, B. yunnanensis, a Chinese species parasitic on Lepidoptera pupa buried in soil that was a sister lineage to B. scarabaeicola.
During surveys of entomopathogenic fungi from different regions in Yunnan Province, China, and Chiang Rai Province, Thailand, over the past 4 years, approximately 15 Beauveria spp. were found and identified (Table 1). In this study, we aimed to: (1) reveal the hidden species diversity of the B. bassiana complex and B. scarabaeidicola complex based on phylogenetic analyses and morphological observation and (2) assess the biocontrol potential of species in the B. bassiana complex and B. scarabaeidicola complex through pathological tests on the lepidopteran Bombyx mori and the coleopteran Tenebrio molitor larvae as well as Protaetia brevitarsis adults.
Materials and Methods
Soil and Specimen Collection
All the soil samples and the majority of Beauveria specimens were collected from Yunnan Province in China. Some specimens were collected from Chiang Rai Province in Thailand. Soil samples and specimens were noted and photographed in the fields, and then carefully placed in plastic containers at low temperature. Afterward, they were carried to the laboratory and stored at 4°C before examination and isolation.
Fungal Isolation and Culture
Beauveria strains were isolated from the soil samples using the Tenebrio molitor baiting method (Keyser et al., 2015). Conidia developing on insect cadavers were transplanted onto plates of potato dextrose agar (PDA; potato 200 g/L, dextrose 20 g/L, agar 20 g/L) and cultured at 25°C. Teleomorph specimens were rinsed with tap water, washed with sterile distilled water, and then dried on sterile filter paper. To obtain axenic cultures, white tissue inside the sclerotia of the teleomorph specimens was removed and inoculated onto PDA plates using a sterilized dissecting knife. Colonies of the isolated filamentous fungi appearing in the culture were transferred onto fresh PDA media. The purified fungal strains were maintained in a culture room at 25°C or transferred to PDA slants and stored at 4°C. Specimens were deposited in the Yunnan Herbal Herbarium (YHH) at the Institute of Herb Biotic Resources of Yunnan University. Cultures were stored in the Yunnan Fungal Culture Collection (YFCC) at the Institute of Herb Biotic Resources of Yunnan University.
Morphological Observations
Specimens were examined using an Olympus SZ61 stereomicroscope (Olympus Corporation, Tokyo, Japan). Cultures on PDA slants were transferred to PDA plates and then incubated at 25°C for 14 days. For morphological evaluation, microscope slides were prepared by placing mycelia from the cultures on PDA medium blocks (5 mm diameter) and then overlaid with a coverslip. Medan dye solution was used to observe asci and ascospores. Other structures were mounted in water. Micro-morphological observations and measurements were performed using a light microscope (CX40, Olympus Corporation, Tokyo, Japan) and a scanning electron microscope (Quanta 200 FEG, FEI Company, Hillsboro, United States). Length to width ratios are given as Q. Mean values for length, width, and Q are indicated by Lm, Wm, and Qm, respectively.
DNA Extraction, PCR and Sequencing
Specimens and axenic living cultures were prepared for DNA extraction. Genomic DNA was extracted using the Genomic DNA Purification Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol. The primer pair nrSSU-CoF and nrSSU-CoR was used to amplify a fraction of the nuclear ribosomal small subunit (nrSSU) (Wang et al., 2015). Primer pair LR5 and LR0R (Vilgalys and Hester, 1990; Rehner and Samuels, 1994) was used to amplify a fraction of the nuclear ribosomal large subunit (nrLSU) and EF1α-EF and EF1α-ER (Bischoff et al., 2006; Sung et al., 2007) for the translation elongation factor 1α (TEF). For amplification of the largest and second largest subunits of the RNA polymerase II (RPB1 and RPB2), PCR primer pairs RPB1-5′F/RPB1-5′R and RPB2-5′F/RPB2-5′R (Bischoff et al., 2006; Sung et al., 2007) were employed. The Bloc fragment was amplified using primer pair B5.1F/B3.1R (Rehner et al., 2006). All the PCR reactions were performed in a final volume of 50 μL and contained 25 μL of 2 × Taq PCR Master Mix (Tiangen Biotech Co. Ltd, Beijing, China), 0.5 μL of each primer (10 μM), 1 μL of genomic DNA, and 23 μL of RNase-free water. Target gene amplification and sequencing were performed according to the methods described in our previous study (Wang Y. B. et al., 2020).
Phylogenetic Analyses
Phylogenetic analyses were based on six gene (nrSSU, nrLSU, TEF, RPB1, RPB2, and Bloc) sequences. The sequences were retrieved from GenBank and combined with those generated in our study. Taxon information and GenBank accession numbers were provided in Supplementary Table 1 and Table 1. Sequences were aligned using MAFFT v.7.1 After alignment, the sequences of the genes were concatenated. Conflicts among the six genes were tested using PAUP* 4.0b10 (Swofford, 2002). The results revealed that the phylogenetic signals in the six genes were not in conflict. The data partitions were defined for the combined dataset using PartitionFinder V1.1.1 (Lanfear et al., 2012). Phylogenetic analyses were conducted using BI and ML methods employing MrBayes v3.1.2 and RaxML 7.0.3, respectively (Ronquist and Huelsenbeck, 2003; Stamatakis et al., 2008). The BI analysis was run on MrBayes v3.1.2 for five million generations using a GTR+G+I model determined by jModelTest version 2.1.4 (Darriba et al., 2012). GTR+I was selected as the optimal model for ML analysis, and 1,000 rapid bootstrap replicates were performed on the dataset.
The first analysis based on the combined five-gene (nrSSU+nrLSU+TEF+RPB1+RPB2) dataset was performed using the following taxa: Akanthomyces, Amphichorda, Ascopolyporus, Beauveria, Blackwellomyces, Cordyceps, Gibellula, Hevansia, Samsoniella, and Simplicillium. Two taxa of Trichoderma were designated as outgroups. The second analysis based on the combined four-gene (TEF+RPB1+RPB2+Bloc) sequences was performed using Beauveria taxa.
We applied a (phylo-) genetic distance matrix calculation for the combined four-gene (TEF+RPB1+RPB2+Bloc) sequences to assess species boundaries in the B. bassiana complex and B. scarabaeidicola complex (Table 2). The pairwise genetic distances of most Beauveria lineages (Supplementary Table 2) were measured based on the Kimura 2-parameter model using MEGA6 software (Tamura et al., 2013).
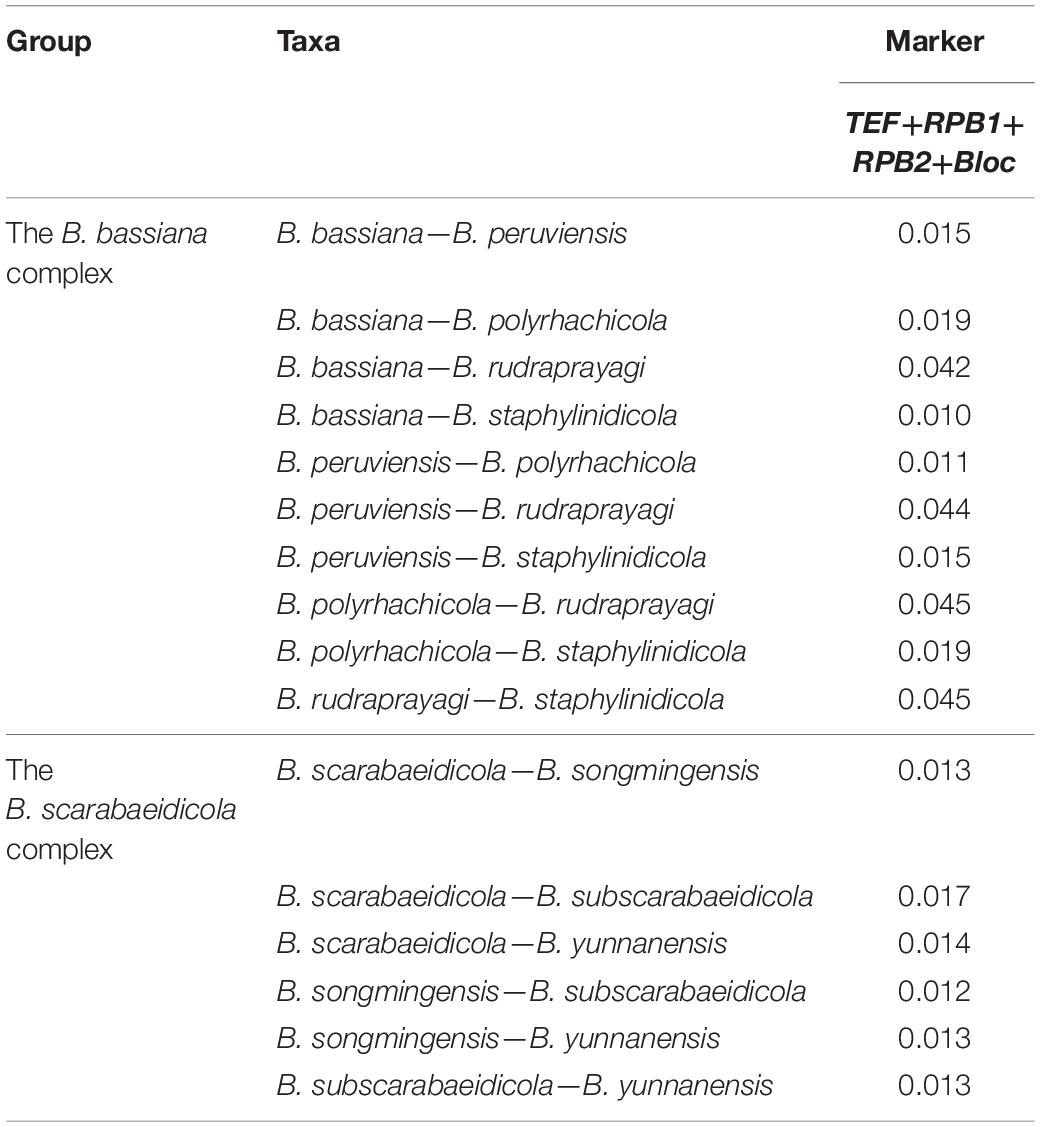
Table 2. Genetic distance (p-distances) of species in the B. bassiana complex and B. scarabaeidicola complex.
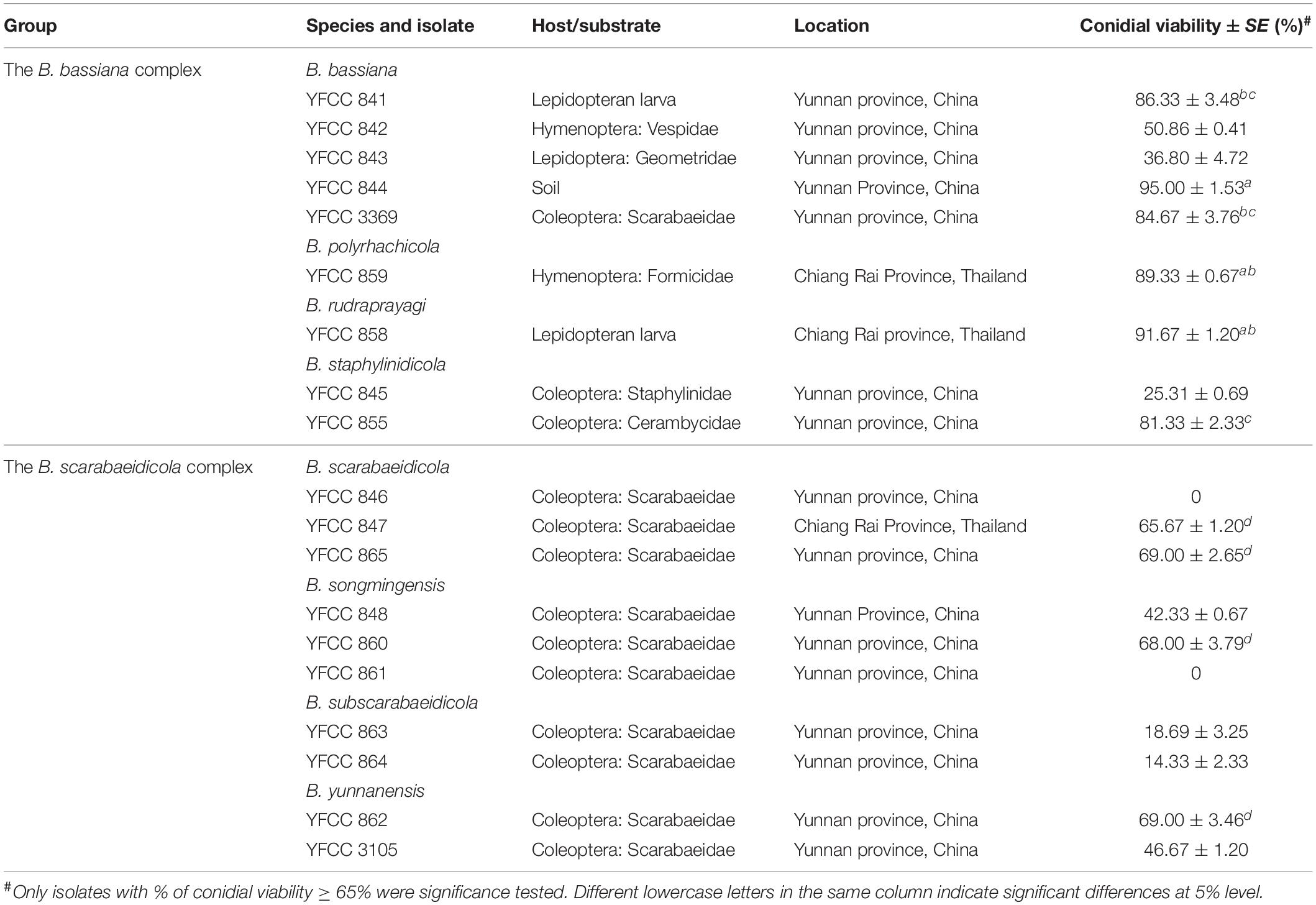
Conidial Viability of Beauveria spp. Isolates
A total of 19 Beauveria spp. isolates (Table 3) were analyzed for their conidial viability using the method described by Imoulan et al. (2011). The conidial viability of each isolate was confirmed by inoculating three tubes of 3 ml PDB media (potato 200 g/L, dextrose 20 g/L) with 0.1 ml of conidia suspension (3 × 106 conidia/ml). Only isolates with conidial viability greater than 65% were tested for pathogenicity toward B. mori, T. molitor, and P. brevitarsis.
Virulence Assay of Beauveria spp. Isolates
A total of 10 Beauveria spp. isolates from the B. bassiana complex and B. scarabaeidicola complex were tested for their pathogenicity to B. mori and T. molitor larvae in addition to P. brevitarsis adults. Conidia for each isolate were obtained from 4-week-old cultures grown on malt extract agar plates, suspended in a sterile aqueous solution of 0.01% Tween 80, and mixed vigorously until homogeneous conidial suspensions were produced. Quantification of the conidia was performed using a hemocytometer under a light microscope at 400 × magnification. All of the suspensions were adjusted to 1 × 108 conidia/ml. The tested insects were individually placed in sterilized rearing boxes and 10 μl of conidial suspension was applied to the surface of each insect. A diet was provided for each insect and renewed as needed. Control groups were treated with the same volume of a sterile aqueous solution of 0.01% Tween 80. The test was replicated three times with 50 insects per replicate. All of the test groups were kept at 25°C under a 12:12 h photoperiod cycle. The numbers of dead insects were recorded every 12 h for a 30 day period, which was used to determine the percentage of mortality. Cadavers were removed, immediately surface-disinfected, and individually placed and maintained in rearing box chambers. Mycelium samples from cadavers were aseptically removed and cultured on PDA for microscope examination, DNA extraction, and TEF sequencing to confirm that mortality was caused by the inoculated fungal strain.
Results
Sequencing and Phylogenetic Analyses
The combined five-gene dataset included sequences from 123 fungal taxa. The final dataset consisted of 5,001 bp of sequence data (nrSSU 1,138 bp, nrLSU 910 bp, TEF 1,047 bp, RPB1 781 bp, and RPB2 1,125 bp). Eleven well-supported clades were recognized based on both Bayesian inference (BI) and maximum likelihood (ML) analyses of the combined five-gene dataset of 123 taxa from Cordycipitaceae and Trichoderma, which accommodate species of the genera Akanthomyces, Amphichorda, Ascopolyporus, Beauveria, Blackwellomyces, Cordyceps, Gibellula, Hevansia, Samsoniella, Simplicillium, and Trichoderma (Supplementary Figure 1). The phylogenetic analyses also revealed the species diversity of the B. bassiana complex and B. scarabaeidicola complex in Beauveria clades. This suggested that the groups composed of the B. bassiana complex and B. scarabaeidicola complex should be genetically composed of at least four species (Supplementary Figure 1). Phylogenetic analyses based on combined partial TEF+RPB1+RPB2+Bloc sequences consisting of 59 fungal taxa resolved most Beauveria lineages in separate terminal branches (Figure 1). This revealed a similar tree and clustering topology, as shown in Supplementary Figure 1. It was proposed that the two strains YFCC 859 and YFCC 867, which formed a strongly supported clade, might be a new species in the B. bassiana complex, named B. polyrhachicola (Figure 1). Our analyses also revealed that two newly discovered species, B. songmingensis and B. subscarabaeidicola, were phylogenetically clustered with B. yunnanensis and B. scarabaeidicola, but they were clearly distinguished from the latter two by forming two separate clades in the B. scarabaeidicola complex (Figure 1 and Supplementary Figure 1). The genetic divergence comparisons showed that the minimum threshold (p-distance) to distinguish genetic species in Beauveria was 0.010 for the combined four-gene (TEF+RPB1+RPB2+Bloc) sequences, as occurred between B. araneola and B. medogensis, between B. asiatica and B. majiangensis, between B. australis and B. brongniartii, and between B. bassiana and B. staphylinidicola (Table 2 and Supplementary Table 2). These results also suggested that: (1) the B. bassiana complex, which is made up of five species, should include B. bassiana, B. peruviensis, B. polyrhachicola, B. rudraprayagi, and B. staphylinidicola and (2) the B. scarabaeidicola complex should be composed of four species, including B. scarabaeidicola, B. songmingensis, B. subscarabaeidicola, and B. yunnanensis.
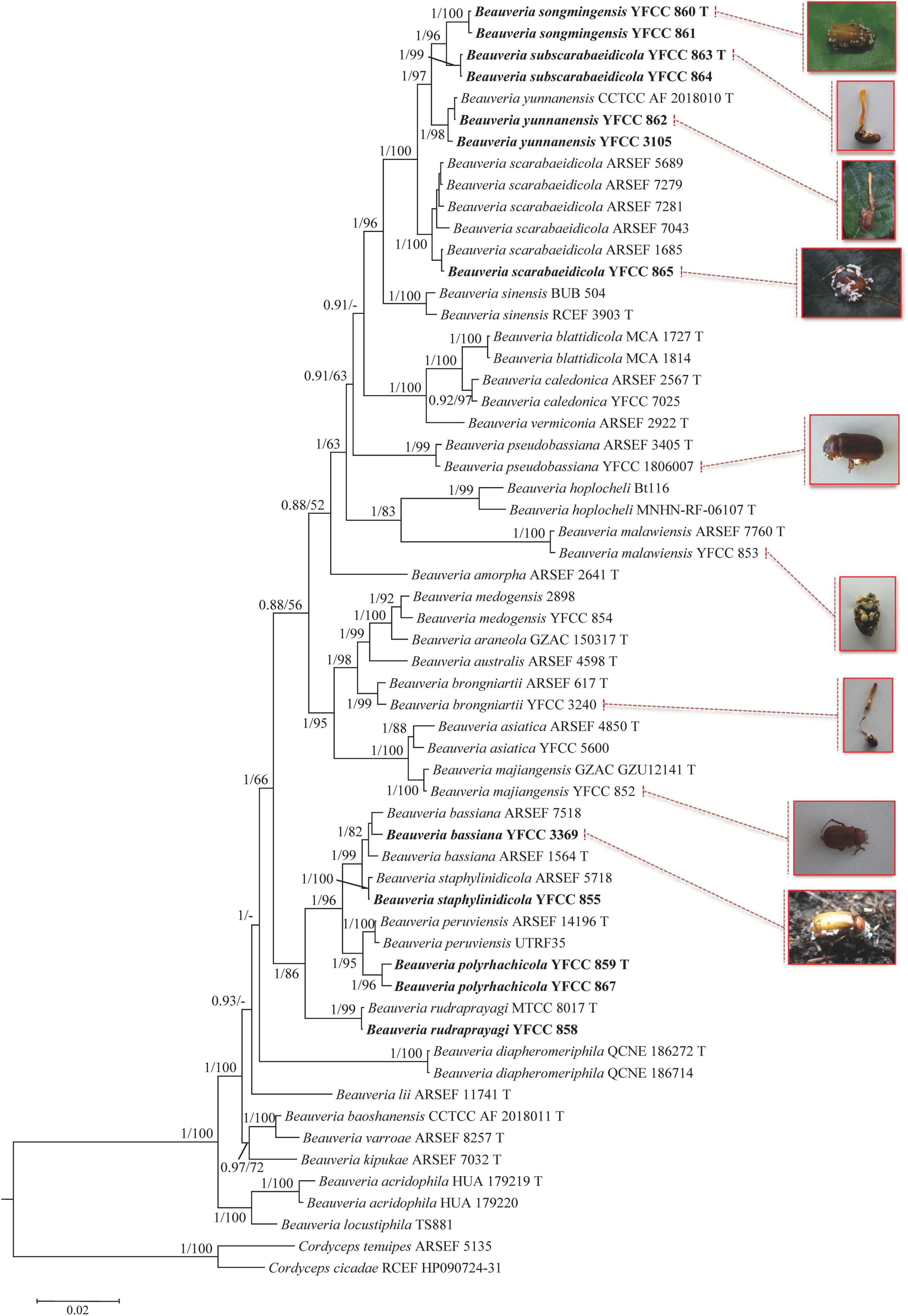
Figure 1. Phylogenetic analysis of Beauveria species based on combined partial TEF+RPB1+RPB2+Bloc sequences. Statistical support values (≥ 0.5/50%) are shown at the nodes for Bayesian inference (BI) posterior probabilities/maximum likelihood (ML) bootstrap support. Isolates representing ex-type material are marked with “T”. Isolates in bold type are those analyzed in this study.
Morphological Features
The morphological characteristics of the three new species as well as photomicrographs of morphological structures are shown in Figures 2–4. The detailed fungal morphological descriptions are provided in the Taxonomy section.

Figure 2. Morphology of Beauveria polyrhachicola. (A,B) The type specimen (YHH 859). (C) Bombyx mori larva infected by B. polyrhachicola. (D) Tenebrio molitor larva infected by B. polyrhachicola. (E) Protaetia brevitarsis adult infected by B. polyrhachicola. (F) Culture character on PDA medium. (G–J) Conidiogenous cells and conidia. Scale bars: (A) = 10 mm; (B) = 2 mm; (C–E) = 10 mm; (F) = 20 mm; (G) = 20 μm; (H–J) = 10 μm.
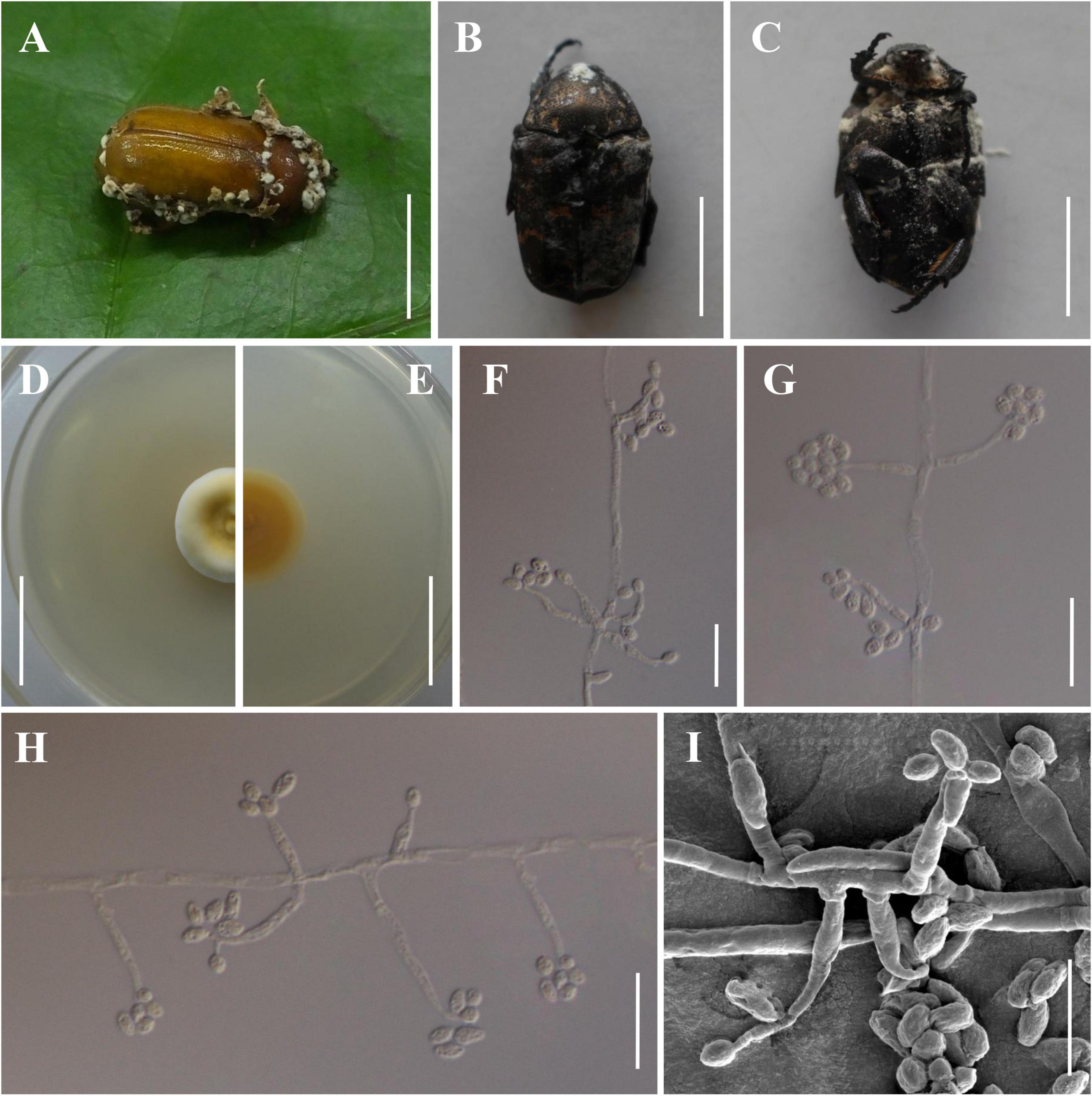
Figure 3. Morphology of Beauveria songmingensis. (A) The type specimen (YHH 860). (B,C) Protaetia brevitarsis adults infected by B. songmingensis. (D,E) Culture character on PDA medium. (F–I) Conidiogenous cells and conidia. Scale bars: (A–C) = 10 mm; (D,E) = 20 mm; (F–H) = 20 μm; (I) = 10 μm.
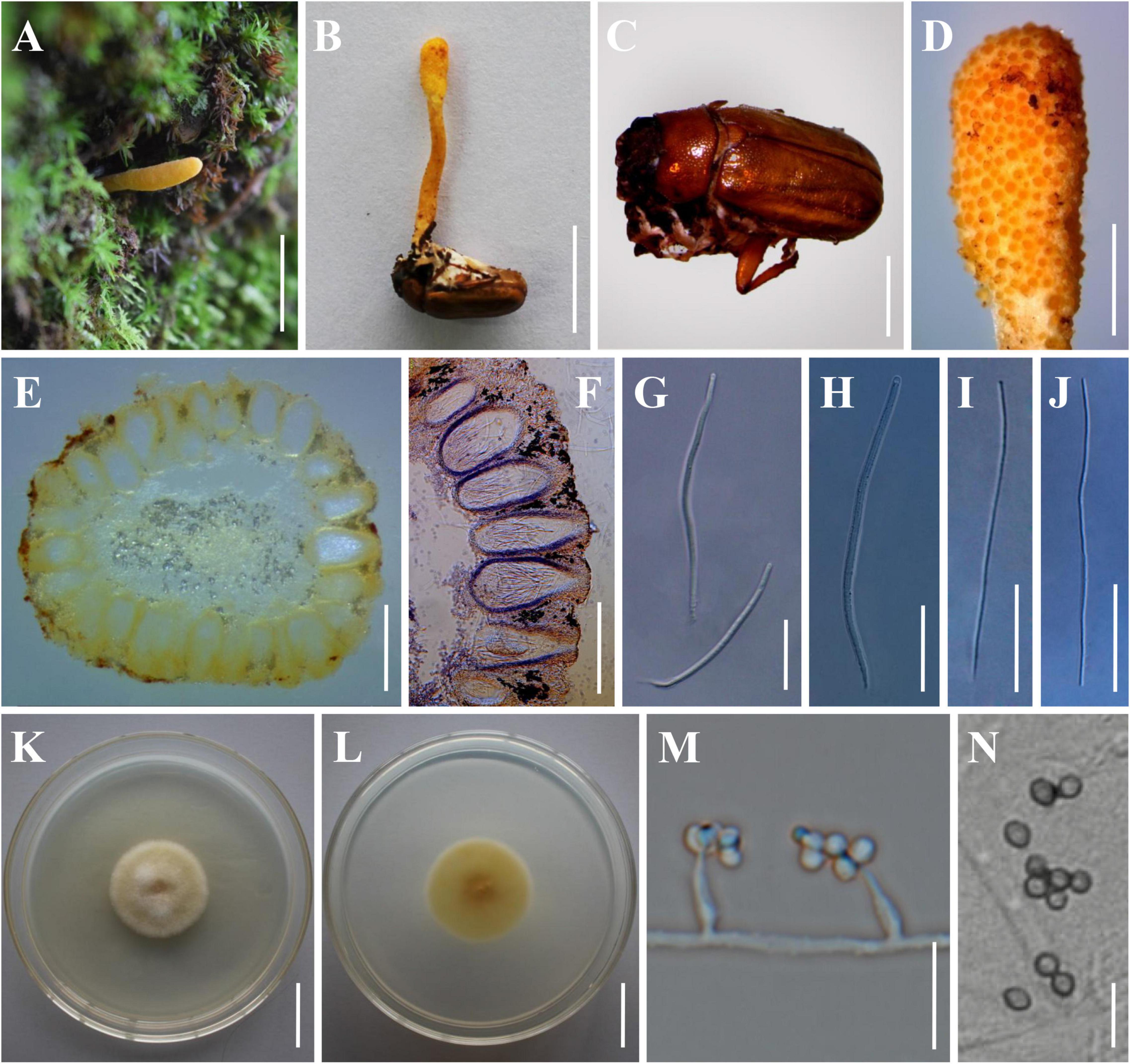
Figure 4. Morphology of Beauveria subscarabaeidicola. (A) Perithecial stroma as encountered in the field. (B) The type specimen (YHH 863). (C) The brown host of B. subscarabaeidicola. (D) Surface of fertile structure of perithecial stroma showing emerging apical parts of immersed perithecia. (E,F) Perithecia. (G,H) Asci. (I,J) Ascospores. (K,L) Culture character on PDA medium. (M,N) Conidiogenous cells and conidia. Scale bars: (A,B) = 10 mm; (C) = 5 mm; (D) = 2 mm; (E,F) = 500 μm; (G–J) = 40 μm; (K,L) = 20 mm; (M,N) = 10 μm.
Conidial Viability of the Beauveria bassiana Complex and Beauveria scarabaeidicola Complex Isolates
Percentage of conidial germination was used to determine conidial viability. The conidial viability of the B. bassiana complex isolates was high, but the highest value ( = 95%) was found on isolates of YFCC 844 from soil (see Table 3). The values of the conidial viability of the isolates in the B. scarabaeidicola complex were significantly lower than those in the B. bassiana complex. Only four B. scarabaeidicola complex isolates with conidial viability were greater than 65%, and their conidial viability values were not significantly different (P < 0.05).
Virulence of the Beauveria bassiana Complex and Beauveria scarabaeidicola Complex Isolates
Ten isolates had conidial viability greater than 65%. These isolates were then selected for pathogenicity tests against B. mori and T. molitor larvae as well as P. brevitarsis adults (Table 4). The B. bassiana complex isolates were shown to have great potential for use in the management of various insect pests; by contrast, the B. scarabaeidicola complex isolates showed obvious host specificity and low virulence. All tested isolates in the B. bassiana complex inflicted mycoses on B. mori and T. molitor larvae and caused over 80% mortality, whereas those in the B. scarabaeidicola complex did not. It was determined that the 10 isolates were pathogenic to P. brevitarsis adults but demonstrated different levels of virulence. Like the conidial viability, the mortalities of P. brevitarsis adults caused by the B. scarabaeidicola complex isolates were significantly lower than those of isolates in the B. bassiana complex (P < 0.05), strengthening the hypothesis that the virulence of certain entomopathogenic fungi is related to their conidial viability (Butt et al., 1994; Fernandes et al., 2007). Additionally, B. bassiana YFCC 844, which was isolated from soil and exhibited the highest conidial viability, showed high virulence against B. mori and T. molitor larvae, as well as P. brevitarsis adults, causing (94.00 ± 1.15)% mortality against B. mori larva, (95.33 ± 1.45)% mortality against T. molitor larva, and (79.00 ± 1.53)% mortality against P. brevitarsis adults (Table 4). Mycelium samples from cadavers were aseptically removed and cultured on PDA. Microscopic examination recovered the same morphological characters of conidiophores and conidia as the inoculated fungal strain. Further, TEF sequenced from DNA extracted from recultures of the external mycelium of cadavers were found to match that of inoculated strain perfectly.
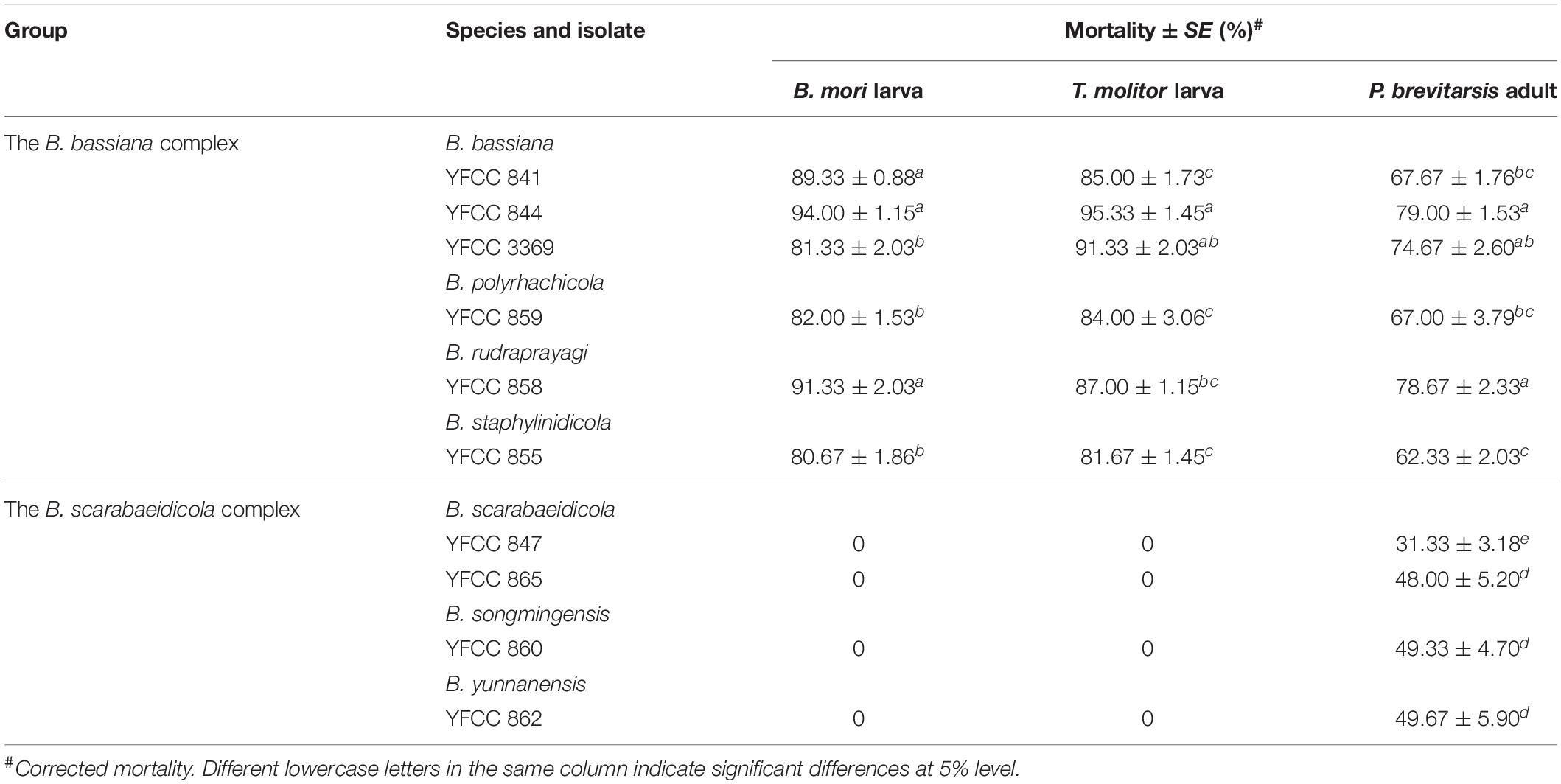
Table 4. The lethal effect of Beauveria spp. isolates on B. mori and T. molitor larvae, and P. brevitarsis adult.
Taxonomy
Beauveria polyrhachicola H. Yu & Y. Wang, sp. nov. Figure 2
MycoBank number 841450.
Etymology: “polyrhachicola” refers to the host (Polyrhachis sp.).
Sexual morph: Undetermined.
Asexual morph: Colonies on PDA reached 20–38 mm in diameter after 14 days at 25°C, white, circular, velutinous, and closely appressed to the agar surface; reverse yellowish white. Odor indistinct. Vegetative hyphae septate, branched, hyaline, smooth-walled, 1.2–2.3 μm wide. Conidiogenous cells, long cylindrical to long flask shaped, solitary or occurring in dense lateral clusters, base cylindrical to ampulliform and 1.4–3.0 μm wide, sympodially branched neck tapering into a long, slender, denticulate rachis, produced laterally on aerial hyphae or from subtending cells, 11.8–40.9 × 1.4–3.0 μm. Conidia 2.0–3.8 × 1.7–2.6 μm, Q = 1.0–1.8 μm (Lm = 2.7 μm, Wm = 2.1 μm, Qm = 1.3), globose, subglobose, slightly ellipsoid, oblong, or cylindrical, hyaline, aseptate, walls smooth and thin.
Type: Thailand, Chiang Rai Province, Khun Tan District (19.9233°N, 100.3133°E, 396 m above sea level), on an adult worker of Polyrhachis sp. emerging from leaf litter on the forest floor, May 2019, collected by Preeyanat Hongthong (holotype: YHH 859; ex-type living culture: YFCC 859).
Distribution: Khun Tan District, Chiang Rai Province, Thailand; Simao District, Yunnan Province, China.
Other material examined: China, Yunnan Province, Puer City, Simao District (22.7113°N, 100.9579°E, 1,360 m above sea level), on an adult worker of Polyrhachis sp. emerging from leaf litter on the forest floor, August 26, 2021, Yao Wang (YHH 867, 868; living culture: YFCC 867).
Notes: Regarding phylogenetic relationships, B. polyrhachicola forms a distinct lineage in the B. bassiana complex, and it is closely related to B. peruviensis, B. staphylinidicola, B. bassiana, and B. rudraprayagi (Figure 1). Morphologically, B. polyrhachicola is similar to B. bassiana, B. kipukae, B. pseudobassiana, B. varroae, and B. peruviensis in terms of the shape and size of the conidia (Rehner et al., 2011; Bustamante et al., 2019). However, B. polyrhachicola can be distinguished from them by its long conidiogenous cells (11.8–40.9 × 1.4–3.0 μm).
Beauveria songmingensis H. Yu & Y. Wang, sp. nov. Figure 3
MycoBank number 841451.
Etymology: named after the location Songming County where this species was collected.
Sexual morph: Undetermined.
Asexual morph: Colonies on PDA reaching 20–35 mm in diameter after 14 days at 25°C, yellowish white, pale yellow, or light yellow, circular; reverse pale yellow, light yellow, or shades of orange to deep orange. Odor indistinct. Vegetative hyphae septate, branched, hyaline or translucent pale yellow, smooth-walled, 2.2–4.5 μm wide. Conidiogenous cells, cylindrical to long flask shaped, solitary but usually in dense clusters of five or more, base cylindrical to ampulliform and 2.7–5.6 μm wide, apex with an indeterminate 1 μm wide geniculate, denticulate rachis, produced laterally on aerial hyphae or from subtending cells, mostly 9.6–34.1 × 2.7–5.6 μm. Conidia 3.6–6.8 × 2.8–3.9 μm, Q = 1.0–2.0 μm (Lm = 5.6 μm, Wm = 3.4 μm, Qm = 1.6), subglobose, broadly ellipsoid, ellipsoid, or oblong, hyaline, aseptate, walls smooth and thin.
Type: China, Yunnan Province, Kunming City, Songming County, Dashao Village (25.3924°N, 102.5589°E, 2,700 m above sea level), on an adult of Pseudosymmachia flavescens (Coleoptera: Scarabaeidae), August 12, 2018, collected by Yao Wang, (holotype: YHH 860; ex-type living culture: YFCC 860).
Distribution: at present known only in Dashao Village, Songming County, Yunnan Province, China.
Other material examined: China, Yunnan Province, Kunming City, Songming County, Dashao Village (25.3924°N, 102.5589°E, 2,700 m above sea level), on an adult of Pseudosymmachia sp. emerging from leaf litter on the forest floor, August 12, 2018, Yao Wang (YHH 848, 861; living culture: YFCC 848, 861).
Notes: Morphologically, B. songmingensis resembles the phylogenetically sister species B. scarabaeidicola and B. subscarabaeidicola. They were found to be parasitic on adult beetles (Coleoptera: Scarabaeidae), and they could be easily recognized by their distinctly yellow colony pigmentation and ellipsoid or oblong conidia. However, our morphological observation revealed a significant difference of conidia sizes between B. songmingensis (3.6–6.8 × 2.8–3.9 μm) and B. scarabaeidicola (2.5–3.5 × 1.5–2.5 μm). B. songmingensis differs from B. subscarabaeidicola by its long conidiogenous cells (9.6–34.1 × 2.7–5.6 μm) and large conidia (3.6–6.8 × 2.8–3.9 μm). Both morphological study and phylogenetic analyses of combined TEF, RPB1, RPB2, and Bloc sequence data support that this fungus is a distinctive species in the genus Beauveria.
Beauveria subscarabaeidicola H. Yu, Y. Wang & Q. Fan, sp. nov. Figure 4
MycoBank number 841452.
Etymology: “subscarabaeidicola” refers to morphologically resembling Beauveria scarabaeidicola but phylogenetically distinct.
Sexual morph: Stromata solitary, fleshy, pale yellow to orange, arising on adult scarab beetles buried in soil or decayed leaves, 30–45 mm long. Stipes cylindrical to clavate, yellowish white to deep yellow, 1.1–2.0 mm wide. Fertile parts clavate, being slightly wider than and indistinct from the stipes, deep yellow to orange, 5.2–26.0 mm long, 1.4–3.3 mm wide. Perithecia semi-immersed and crowded at the apex of the stromata, ampuliform, pyriform, ovoid to oblong, 265–700 × 180–320 μm (n = 50). Asci hyaline, cylindrical, 124.5–257.4 × 3.7–5.2 μm (n = 50). Apical caps prominent, hemiglobose, 2.7–3.9 μm wide, 2.4–3.2 μm high (n = 50). Ascospores hyaline, filiform, multi-septate, finally breaking into secondary ascospores, 75.6–188.5 × 1.0–1.5 μm (n = 30). Secondary ascospores cylindrical, hyaline, 6.9–11.2 × 1.0–1.5 μm (n = 50).
Asexual morph: Colonies on PDA reaching 28–42 mm in diameter after 14 days at 25°C, yellowish white, pale yellow, or light yellow, circular; reverse pale yellow, light yellow, or shades of orange to deep orange. Odor indistinct. Vegetative hyphae septate, branched, hyaline or translucent pale yellow, smooth-walled, 1.2–2.5 μm wide. Conidiogenous cells, phialidic, solitary but usually in dense clusters of five or more, base subspherical to ampulliform and 2.8–5.0 μm wide, apex with an indeterminate 1 μm wide geniculate, denticulate rachis, produced laterally on aerial hyphae or from subtending cells mostly 4.8–6.9 × 2.0–4.6 μm. Conidia 2.6–4.2 × 1.9–3.5 μm, Q = 1.0–1.4 μm (Lm = 3.4 μm, Wm = 2.8 μm, Qm = 1.2), subglobose or broadly ellipsoid, hyaline, aseptate, walls smooth and thin.
Type: China, Yunnan Province, Kunming City, Songming County, Dashao Village (25.2398°N, 102.5617°E, 2,697 m above sea level), on an adult of Anomala exoleta (Coleoptera: Scarabaeidae), July 23rd, 2019, collected by Dexiang Tang, (holotype: YHH 863; ex-type living culture: YFCC 863).
Distribution: at present known only from Dashao Village, Songming County, Yunnan Province, China.
Other material examined: China, Yunnan Province, Kunming City, Songming County, Dashao Village (25.2398°N, 102.5617°E, 2,697 m above sea level), on an adult of Anomala exoleta, July 23, 2019, Dexiang Tang (YHH 864; living culture: YFCC 864).
Notes: Beauveria subscarabaeidicola is practically indistinguishable in morphology to B. scarabaeidicola. Our morphological observation revealed no significant differences in the morphological characteristics of teleomorph and anamorph between the two species (Kobayasi and Shimizu, 1976; Rehner et al., 2011; Shrestha et al., 2014). The lack of diagnostic morphological features to distinguish B. subscarabaeidicola and B. scarabaeidicola was overcome by delimiting the two species using DNA-based methodologies.
Discussion
It is generally agreed that distinguishing individual Beauveria species can be difficult using only morphological characters, as several species in the genus are morphologically cryptic species. In this study, we conducted a comprehensive investigation of the cryptic species diversity of the B. bassiana complex and B. scarabaeidicola complex. The molecular phylogeny clearly suggested the existence of distinct species in the B. bassiana complex and B. scarabaeidicola complex that we accordingly propose as new species: B. polyrhachicola (Figure 2), B. songmingensis (Figure 3), and B. subscarabaeidicola (Figure 4). Beauveria polyrhachicola is practically indistinguishable in morphology from other members of the B. bassiana complex. The shape and size of the conidia and the colony color of B. polyrhachicola, among other morphological features, have been observed in B. bassiana, B. rudraprayagi, B. staphylinidicola, and B. peruviensis (Rehner et al., 2011; Agrawal et al., 2014; Kepler et al., 2017; Bustamante et al., 2019). In the B. scarabaeidicola complex, the macromorphology of B. scarabaeidicola, B. songmingensis, and B. subscarabaeidicola is very similar, and thus species cannot be distinguished visually. The macroscopic and microscopic observations performed during our investigation revealed the extensive overlap in morphological characters and the lack of distinctive phenotypic variation, supporting the notion of cryptic species in a species complex.
At present, multi-locus phylogenetic analyses have gained importance in delimiting the species within the entomopathogenic fungi Beauveria. Rehner et al. (2011) divided B. bassiana s. lat. and B. brongniartii s. lat. into several cryptic species and described six new species based on the Bloc nuclear intergenic region and three nuclear genes encoding elongation factor 1-a (TEF), RNA polymerase II largest subunit (RPB1), and RNA polymerase II second largest subunit (RPB2). Subsequently, more than seven new species and new combinations were confirmed using combined analysis of the four-locus sequence data (Zhang et al., 2012; Chen et al., 2013, 2017, 2018; Agrawal et al., 2014; Robène-Soustrade et al., 2015; Imoulan et al., 2016). In more recent studies, six species were added to the genus based on multilocus (nrSSU, nrLSU, TEF, RPB1, and RPB2) sequence data: B. acridophila, B. blattidicola, B. diapheromeriphila, B. locustiphila, B. scarabaeidicola, and B. staphylinidicola (Kepler et al., 2017). In this study, we analyzed most species of the newly circumscribed genus Beauveria based on phylogenetic inferences of six nuclear molecular markers (nrSSU, nrLSU, TEF, RPB1, RPB2, and Bloc). Phylogenetic analyses based on the five-gene (nrSSU, nrLSU, TEF, RPB1, and RPB2) dataset and the combined four-gene (TEF+RPB1+RPB2+Bloc) sequences produced trees with similar topologies that resolved most Beauveria lineages in separate terminal branches (Figure 1 and Supplementary Figure 1). The results of the present work indicate that the first dataset was conducive to determining the phylogenomic relationships between Beauveria and its related genera, and the use of the latter was essential to establish robust Beauveria species boundaries, particularly the B. bassiana complex and B. scarabaeidicola complex.
Scarab beetles are leaf and root feeding pests of grasses, grains, sugarcane, strawberry, potato tubers, and young nursery plants (Crocker et al., 1996; Yokoyama et al., 1998). Based on the published literature, there are about six Beauveria spp. that parasitize adult scarab beetles: B. asiatica, B. bassiana, B. brongniartii, B. majiangensis, B. pseudobassiana, and B. scarabaeidicola (Rehner et al., 2011; Kepler et al., 2017; Chen et al., 2018; Khonsanit et al., 2020; Wang Y. et al., 2020). Here, we identified an extension of the members to also include B. malawiensis, B. songmingensis, B. subscarabaeidicola, and B. yunnanensis, as shown in Figure 1. Chen et al. (2019) emphasized that hosts of B. yunnanensis isolates were Lepidoptera pupae. However, our morphological observations of specimens from a type locality of B. yunnanensis indicated that their hosts were adult scarab beetles. Moreover, the host of B. yunnanensis was not shown in their publication (Chen et al., 2019). It seems that the host of Lepidoptera pupa is doubtful. There is reason to believe that members of the B. scarabaeidicola species complex are host-specific.
Not all scarab-killing pathogens are suitable for mycoinsecticide formulations that control scarab beetles. Our data suggested that the B. scarabaeidicola complex isolates showed low virulence. In addition, mortalities in P. brevitarsis adults caused by the B. scarabaeidicola complex isolates were significantly lower than those of isolates in the B. bassiana complex. Additional research is needed to determine the effectiveness of other species before future consideration of isolates for biological pest control.
Conclusion
The B. bassiana complex and B. scarabaeidicola complex, as special groups in the genus Beauveria, are rich in species diversity and have a wide distribution in nature. The B. bassiana complex, which is made up of five species, is a cosmopolitan group of soilborne necrotrophic arthropod-pathogenic fungi that have been shown to have great potential for the management of various insect pests. The B. scarabaeidicola complex is composed of pathogens specific to scarab beetles, and it is found on leaf litter or buried in soil. Species in this complex are morphologically highly similar and can hardly be distinguished macroscopically. In this study, we reported the discovery and description of three new species: B. polyrhachicola, which was found in the B. bassiana complex, and B. songmingensis and B. subscarabaeidicola, which were found in the B. scarabaeidicola complex. In addition, 10 species of Beauveria were found to be parasitic on scarab beetles. However, not all members are suitable for mycoinsecticide formulations for controlling scarab beetles. Our data suggested that the B. scarabaeidicola complex isolates showed obvious low virulence. Additionally, the mortality of Protaetia brevitarsis adults caused by the B. scarabaeidicola complex isolates was significantly lower than that of isolates in the B. bassiana complex.
Data Availability Statement
The datasets presented in this study can be found in GenBank. The accession numbers can be found in the article/Supplementary Material.
Author Contributions
YW and HY: conceptualization. YW: methodology, writing—original draft preparation, and formal analysis. YW and QF: software. QF, W-QZ, and DW: validation. YW, DW, D-XT, PH, and HY: investigation. YW, D-XT, and PH: resources. HY: writing—review and editing and funding acquisition. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31870017, 31760011, and 32160005) and the Department of Science and Technology of Yunnan Province [No. 2018FY001(-006)].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Yuan-Bing Wang for help in naming the new fungal species.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.841604/full#supplementary-material
Footnotes
References
Agrawal, Y., Mual, P., and Shenoy, B. D. (2014). Multi-gene genealogies reveal cryptic species Beauveria rudraprayagi sp. nov. from India. Mycosphere 5, 719–736. doi: 10.5943/mycosphere/5/6/3
Bischoff, J. F., Rehner, S. A., and Humber, R. A. (2006). Metarhizium frigidum sp. nov.: a cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia 98, 737–745. doi: 10.1080/15572536.2006.11832645
Bustamante, D. E., Oliva, M., Leiva, S., Mendoza, J. E., Bobadilla, L., Angulo, G., et al. (2019). Phylogeny and species delimitations in the entomopathogenic genus Beauveria (Hypocreales, Ascomycota), including the description of B. peruviensis sp. nov. MycoKeys 58, 47–68. doi: 10.3897/mycokeys.58.35764
Butt, T. M., Ibrahim, L., Ball, B. V., and Clark, S. J. (1994). Pathogenicity of the entomogenous fungi Metarhizium anisopliae and Beauveria bassiana against crucifer pests and the honey bee. Biocontrol Sci. Technol. 4, 207–214. doi: 10.1080/09583159409355328
Chen, M. J., Huang, B., Li, Z. Z., and Spatafora, J. W. (2013). Morphological and genetic characterisation of Beauveria sinensis sp. nov. from China. Mycotaxon 124, 301–308. doi: 10.5248/124.301
Chen, W. H., Han, Y. F., Liang, Z. Q., and Jin, D. C. (2017). A new araneogenous fungus in the genus Beauveria from Guizhou, China. Phytotaxa 302, 57–64. doi: 10.11646/phytotaxa.302.1.5
Chen, W. H., Liu, M., Huang, Z. X., Yang, G. M., Han, Y. F., Liang, J. D., et al. (2018). Beauveria majiangensis, a new entomopathogenic fungus from Guizhou, China. Phytotaxa 333, 243–250. doi: 10.11646/phytotaxa.333.2.8
Chen, Z. H., Chen, K., Dai, Y. D., Zheng, Y., Wang, Y. B., Yang, X. N., et al. (2019). Beauveria species diversity in the Gaoligong Mountains of China. Mycol. Prog. 18, 933–943. doi: 10.1007/s11557-019-01497-z
Crocker, R. L., Rodriguez-del-Bosque, L. A., Nailon, W. T., and Wei, X. (1996). Flight periods in Texas of three parasites (Diptera: Pyrgotidae) of adult Phyllophaga spp. (Coleoptera: Scarabaeidae) and egg production by Pyrgota undata. Southwest. Entomol. 21, 317–324.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Faria, M. R., and Wraight, S. P. (2007). Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43, 237–256. doi: 10.1016/j.biocontrol.2007.08.001
Fernandes, ÉK. K., Rangel, D. E. N., Moraes, ÁM. L., Bittencourt, V. R. E. P., and Roberts, D. W. (2007). Variability in tolerance to UV-B radiation among Beauveria spp. isolates. J. Invertebr. Pathol. 96, 237–243. doi: 10.1016/j.jip.2007.05.007
Goettel, S., Eilenberg, J., and Glare, T. (2005). “Entomopathogenic fungi and their role in regulation of insect populations,” in Comprehensive Molecular Insect Science, eds L. B. Gilbert and K. Latrou (Oxford: Elsevier), 361–406.
Imoulan, A., Alaoui, A., and El Meziane, A. (2011). Natural occurrence of soil-borne entomopathogenic fungi in the Moroccan endemic forest of Argania spinosa and their pathogenicity to Ceratitis capitata. World J. Microbiol. Biotechnol. 27, 2619–2628. doi: 10.1007/s11274-011-0735-1
Imoulan, A., Wu, H. J., Lu, W. L., Li, Y., Li, B. B., Yang, R. H., et al. (2016). Beauveria medogensis sp. nov., a new fungus of the entomopathogenic genus from China. J. Invertebr. Pathol. 139, 74–81. doi: 10.1016/j.jip.2016.07.006
Kepler, R. M., Luangsa-ard, J. J., Hywel-Jones, N. L., Quandt, C. A., Sung, G. H., Rehner, S. A., et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8, 335–353. doi: 10.5598/imafungus.2017.08.02.08
Kepler, R. M., Sung, G. H., Ban, S., Nakagiri, A., Chen, M. J., Huang, B., et al. (2012). New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 104, 182–197. doi: 10.3852/11-070
Keyser, C. A., De Fine Licht, H. H., Steinwender, B. M., and Meyling, N. V. (2015). Diversity within the entomopathogenic fungal species Metarhizium flavoviride associated with agricultural crops in Denmark. BMC Microbiol. 15:249. doi: 10.1186/s12866-015-0589-z
Khonsanit, A., Luangsa-ard, J. J., Thanakitpipattana, D., Noisripoom, W., Chaitika, T., and Kobmoo, N. (2020). Cryptic diversity of the genus Beauveria with a new species from Thailand. Mycol. Prog. 19, 291–315. doi: 10.1007/s11557-020-01557-9
Kobayasi, Y., and Shimizu, D. (1976). The genus Cordyceps and its allies from New Guinea. Bull. Natl. Sci. Mus. Ser. B 2, 133–151.
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Li, Z. Z., Huang, B., Chen, M. J., Wang, B., and Fan, M. Z. (2011). Studies on the genus Beauveria in molecular era. Mycosystema 30, 823–835.
Luo, Z., Zhang, T., Liu, P., Bai, Y., Chen, Q., Zhang, Y., et al. (2018). The Beauveria bassiana Gas 3 β-glucanosyltransferase contributes to fungal adaptation to extreme alkaline conditions. Appl. Environ. Microbiol. 84:e01086-18. doi: 10.1128/AEM.01086-18
McKinnon, A. C., Glare, T. R., Ridgway, H. J., Mendoza-Mendoza, A., Holyoake, A., Godsoe, W. K., et al. (2018). Detection of the entomopathogenic fungus Beauveria bassiana in the rhizosphere of wound-stressed Zea mays plants. Front. Microbiol. 9:1161. doi: 10.3389/fmicb.2018.01161
Ownley, B. H., Gwinn, K. D., and Vega, F. E. (2010). Endophytic fungal entomopathogens with activity against plant pathogens: ecology and evolution. Biocontrol 55, 113–128. doi: 10.1007/s10526-009-9241-x
Rehner, S. A., Minnis, A. M., Sung, G. H., Luangsa-ard, J. J., Devotto, L., and Humber, R. A. (2011). Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103, 1055–1073. doi: 10.3852/10-302
Rehner, S. A., Posada, F., Buckley, E. P., Infante, F., Castillo, A., and Vega, F. E. (2006). Phylogenetic origins of African and Neotropical Beauveria bassiana s.l. pathogens of the coffee berry borer, Hypothenemus hampei. J. Invertebr. Pathol. 93, 11–21. doi: 10.1016/j.jip.2006.04.005
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/S0953-7562(09)80409-7
Robène-Soustrade, I., Jouen, E., Pastou, D., Payet-Hoarau, M., Goble, T., Linderme, D., et al. (2015). Description and phylogenetic placement of Beauveria hoplocheli sp. nov. used in the biological control of the sugarcane white grub, Hoplochelus marginalis, on Reunion Island. Mycologia 107, 1221–1232. doi: 10.3852/14-344
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sanjuan, T., Tabima, J., Restrepo, S., Læssøe, T., Spatafora, J. W., and Franco-Molano, A. E. (2014). Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordyceps sensu stricto). Mycologia 106, 260–275. doi: 10.3852/13-020
Shrestha, B., Hyun, M. W., Oh, J., Han, J. G., Lee, T. H., Cho, J. Y., et al. (2014). Molecular evidence of a teleomorph-anamorph connection between Cordyceps scarabaeicola and Beauveria sungii and its implication for the systematics of Cordyceps sensu stricto. Mycoscience 55, 231–239. doi: 10.1016/j.myc.2013.09.004
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Sung, G. H., Hywel-Jones, N. L., Sung, J. M., Luangsa-ard, J. J., Shrestha, B., and Spatafora, J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.57.01
Swofford, D. L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sunderland, MA: Sinauer.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cyptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, Y., Tang, D. X., Duan, D. E., Wang, Y. B., and Yu, H. (2020). Morphology, molecular characterization, and virulence of Beauveria pseudobassiana isolated from different hosts. J. Invertebr. Pathol. 172:107333. doi: 10.1016/j.jip.2020.107333
Wang, Y. B., Wang, Y., Fan, Q., Duan, D. E., Zhang, G. D., Dai, R. Q., et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 103, 1–46. doi: 10.1007/s13225-020-00457-3
Wang, Y. B., Yu, H., Dai, Y. D., Wu, C. K., Zeng, W. B., Yuan, F., et al. (2015). Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps sp. infecting melolonthid larvae in southwestern China. Mycol. Prog. 14:70. doi: 10.1007/s11557-015-1090-7
Yokoyama, T., Hasegawa, M., Fujiie, A., Sawada, M., and Noguchi, K. (1998). Microbial control of scarab beetle larvae by a formulation of Metarhizium anisopliae (Deuteromycotina: Hyphomycetes) in a sweet potato field. Appl. Entomol. Zool. 33, 215–218. doi: 10.1303/aez.33.215
Zhang, S. L., He, L. M., Chen, X., and Huang, B. (2012). Beauveria lii sp. nov. isolated from Henosepilachna vigintioctopunctata. Mycotaxon 121, 199–206. doi: 10.5248/121.199
Keywords: Taxonomy, Beauveria, phylogenetic analyses, new species, biological control
Citation: Wang Y, Fan Q, Wang D, Zou W-Q, Tang D-X, Hongthong P and Yu H (2022) Species Diversity and Virulence Potential of the Beauveria bassiana Complex and Beauveria scarabaeidicola Complex. Front. Microbiol. 13:841604. doi: 10.3389/fmicb.2022.841604
Received: 22 December 2021; Accepted: 14 February 2022;
Published: 04 March 2022.
Edited by:
Dimitrios Georgios Karpouzas, University of Thessaly, GreeceReviewed by:
Andreas Leclerque, Darmstadt University of Technology, GermanySamantha Chandranath Karunarathna, Qujing Normal University, China
Copyright © 2022 Wang, Fan, Wang, Zou, Tang, Hongthong and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Yu, hongyu@ynu.edu.cn
 Yao Wang1,2
Yao Wang1,2 Hong Yu
Hong Yu