- 1Center for Mycomedicine Research, Basic Medical School, Guizhou University of Traditional Chinese Medicine, Guiyang, China
- 2Institute of Fungus Resources, Department of Ecology, College of Life Sciences, Guizhou University, Guiyang, China
Simplicillium species are widely distributed and commonly found on various substrates. A minority of species are associated with arthropods. A spider-associated species Simplicillium araneae, and three insect-associated species, Simplicillium coleopterorum, Simplicillium guizhouense, and Simplicillium larvatum, are proposed as novel species based on a multi-locus phylogenetic analysis and morphological characteristics. These Simplicillium species completely fit the nutritional model of Hypocreales fungi and could be used as a model to study their evolutionary relationship. A phylogenetic network analysis based on ITS sequences suggests that a host jump was common among Simplicillium species, and S. araneae may have originally come from an insect host and then jumped to a spider host. However, the evolutionary relationship of S. coleopterorum, S. guizhouense, and S. larvatum was not clear in the phylogenetic network and more sequencing information should be added to the network. In addition, strain CBS 101267 was identified as Simplicillium subtropicum.
Introduction
The genus Simplicillium branched off from the genus Verticillium section Prostrata, and it consists of four species: S. lanosoniveum (J.F.H. Beyma) Zare and W. Gams, S. obclavatum (W. Gams) Zare and W. Gams, S. lamellicola (F.E.V. Sm.) Zare and W. Gams, and S. wallacei H.C. Evans (Zare and Gams, 2001). Zare and Gams (2001) summarized that solitary phialides, conidia adhering in globose, slimy heads or imbricate chains, and crystals commonly present in agar were the typical characteristics of Simplicillium. After that, numerous species were added to the genus (Liu and Cai, 2012; Nonaka et al., 2013; Gams, 2017; Zhang et al., 2017; Crous et al., 2018; Gomes et al., 2018; Chen et al., 2019, 2021; Wei et al., 2019; Kondo et al., 2020; Wang et al., 2020; Leplat et al., 2021). However, based mainly on rDNA sequence analyses, several Simplicillium species (S. wallacei, S. coffeanum A.A.M. Gomes and O.L. Pereira, S. chinensis F. Liu and L. Cai, and S. filiforme R.M.F. Silva, R.J.V. Oliveira, Souza-Motta, J.L. Bezerra and G.A. Silva) were transferred to the genera Lecanicillium W. Gams and Zare and Leptobacillium Zare and W. Gams (Zare and Gams, 2008; Okane et al., 2020; Chen et al., 2021). As a result, the genus Simplicillium currently consists of 23 species.
Chen et al. (2021) noted that Simplicillium species inhabit diverse substrates and could be used as a model of Hypocreales fungi to study their evolutional relationship. However, the phylogenetic tree assumes that biological groups evolve in the form of tree divergence and cannot accurately present the whole process of actual evolution, including hybridization, horizontal gene transfer, and gene recombination within the population (Cheng and Huang, 2008). The neighbor-net network (split network), a kind of distance-based phylogenetic network, can be used to present conflicting and ambiguous signals in datasets and detect subtle differences (Bryant and Moulton, 2004; Huson and Bryant, 2006). It can provide a way to present parallel events that are covered up and cannot be displayed by a phylogenetic tree, as well as an uncertain evolutionary phylogenetic relationship. The method has been applied in the phylogenetic analysis of animals, plants, and microorganisms (Bandelt and Dress, 1992; Morrison, 2005; Huson and Bryant, 2006; Morozov et al., 2013; Khonsanit et al., 2020).
During a survey of entomopathogenic fungi from Southwest China, some insect- and spider-associated specimens were found and some new Simplicillium strains were isolated and purified. The goals of this research were as follows: (1) identify the new strains based on ITS sequence, (2) characterize the new species of the genus Simplicillium based on a multi-locus phylogenetic analysis and their morphological and ecological characteristics, and (3) detect the evolutional relationship of the new species by the neighbor-net network based on ITS sequence of Simplicillium species.
Materials and methods
Specimen collection and identification
Five infected insect and spider specimens (DY1005, DY1025, DY10173, DY10181, and SD0538) were collected from Duyun City (26°21'24.71” N, 107°22'48.22” E) and Sandu County (25°57'22.21” N, 107°57'54.69” E), Guizhou Province, on 1 October and 1 May 2019. The surface of each insect body was rinsed with sterile water, followed by surface sterilization with 75% ethanol for 3–5 s and rinsing 3 times with sterilized water. After drying on sterilized filter paper, the synnemata, mycelium, or a part of the sclerotia was removed from the specimen, inoculated on potato dextrose agar (PDA), and improved potato dextrose agar (PDA, 1% w/v peptone) plates (Chen et al., 2019). Fungal colonies emerging from the specimens were isolated and cultured at 25°C for 14 days under 12 h light/12 h dark conditions following protocols described by Zou et al. (2010). The specimens and isolated strains were deposited at the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China.
Macroscopic characterization was determined from PDA cultures incubated at 25°C for 14 days, and the growth rate of the colony, the presence of octahedral crystals, and the colony colors (surface and reverse) were observed. To investigate the microscopic characteristics, a small number of mycelia were mounted in lactophenol cotton blue or 20% lactate acid solution and observed with an optical microscope (OM, DM4 B, Leica, Germany).
DNA extraction, polymerase chain reaction amplification, and nucleotide sequencing
DNA extraction was carried out using a fungal genomic DNA extraction kit (DP2033, BioTeke Corporation) according to Liang et al. (2011). The extracted DNA was stored at −20°C. Amplification of the internal transcribed spacer (ITS) region, large subunit ribosomal RNA (LSU) gene, small subunit ribosomal RNA (SSU), RNA polymerase II largest subunit 1 (RPB1), and translation elongation factor 1 alpha (TEF) was carried out by PCR as described by White et al. (1990), Rakotonirainy et al. (1994), and Castlebury et al. (2004). Primer sequence information is shown in Supplementary Table S1. PCR products were purified and sequenced at Sangon Biotech (Shanghai) Co. The resulting sequences were submitted to GenBank (Table 1).
Sequence alignment and phylogenetic and network analyses
Lasergene software (version 6.0, DNASTAR) was used to edit DNA sequences in this study. The SSU, ITS, LSU, RPB1, and TEF sequences were downloaded from GenBank, based on Nonaka et al. (2013), Zhang et al. (2017), Crous et al. (2018, 2021), Gomes et al. (2018), Chen et al. (2019, 2021), Wei et al. (2019), Kondo et al. (2020), Leplat et al. (2021), and others selected based on BLAST searches in GenBank (Table 1). ITS sequence was applied to identify the new strains in the genus Simplicillium (analysis 1). ITS sequences and other loci were aligned and edited by MAFFT v7.037b (Katoh and Standley, 2013) and MEGA6 (Tamura et al., 2013). Combined sequences of SSU, ITS, LSU, RPB1, and TEF were applied to establish the four novel species (analysis 2) and obtained using SequenceMatrix v.1.7.8 (Vaidya et al., 2011). The model was selected for Bayesian analysis by ModelFinder (Kalyaanamoorthy et al., 2017) in PhyloSuite software Zhang et al. (2020a).
ITS sequences and the combined loci were analyzed using Bayesian inference (BI) and maximum likelihood (ML) methods. For BI, a Markov chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.2 (Ronquist et al., 2012) for the combined sequence datasets. The Bayesian analysis resulted in 20,001 trees after 10,000,000 generations. The first 4,000 trees, representing the burn-in phase of the analysis, were discarded, while the remaining 16,001 trees were used to calculate posterior probabilities in the majority rule consensus tree. After the analysis was finished, each run was examined using the program Tracer v1.5 (Drummond and Rambaut, 2007) to determine burn-in and confirm that both runs had converged. ML analyses were constructed with RAxMLGUI (Silvestro and Michalak, 2012). The GTRGAMMA model was used for all partitions, in accordance with recommendations in the RAxML manual against the use of invariant sites. The phylogenetic network was constructed by SplitsTree 4 using the neighbor-net method based on the ITS sequence, which was the most common sequence in the genus Simplicillium. The other condition was the default settings (Huson and Bryant, 2006).
Results
Phylogenetic and network analyses
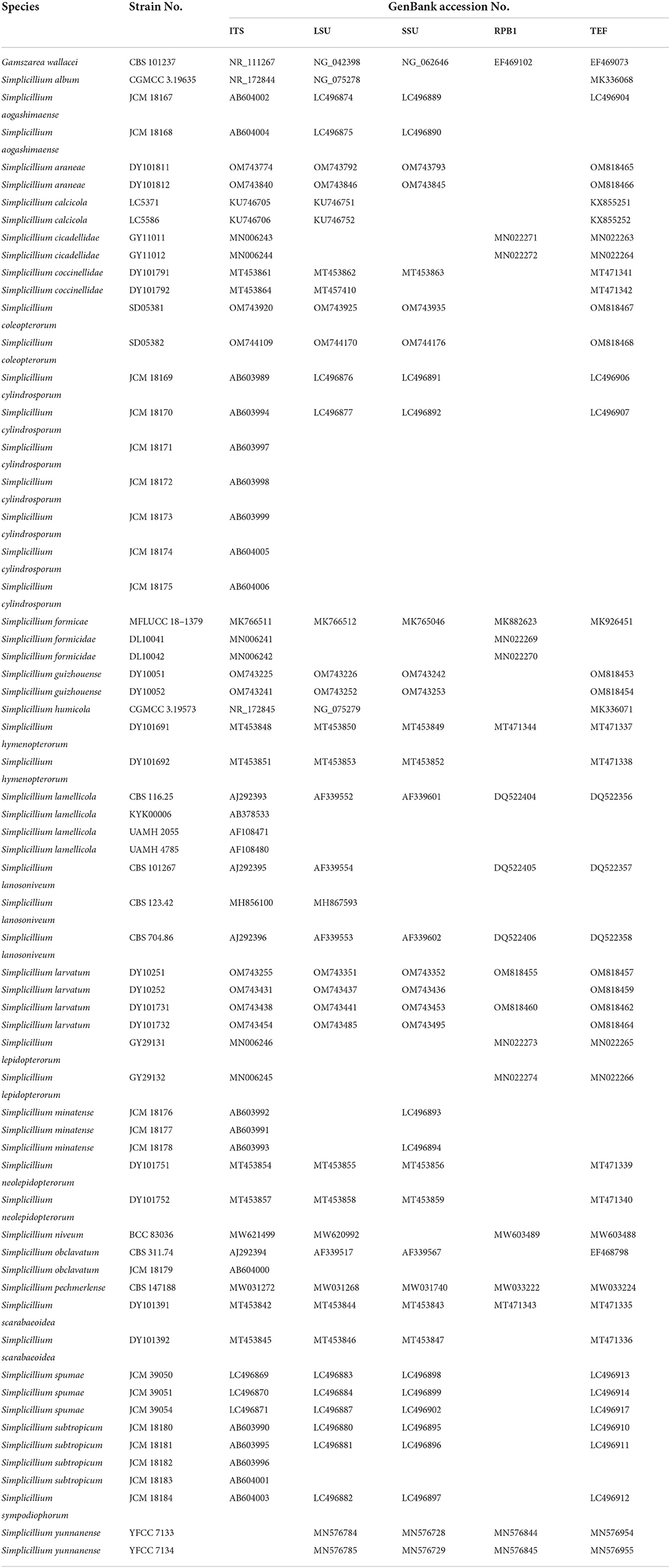
In the phylogenetic tree of analysis 1 (to identify new strains in the genus Simplicillium) and analysis 2 (to establish four novel species) (Figures 1, 2, respectively), Gamszarea wallacei (H.C. Evans) Z.F. Zhang and L. Cai (CBS 101237) was used as the outgroup. The ITS sequence and concatenated sequences (SSU, ITS, LSU, RPB1, and TEF) of analyses 1 and 2 included 23 and 24 taxa, respectively, and consisted of 547 (ITS) and 3,544 (SSU, 982; ITS, 547; LSU, 624; RPB1, 544; and TEF, 847) characters with gaps, respectively.
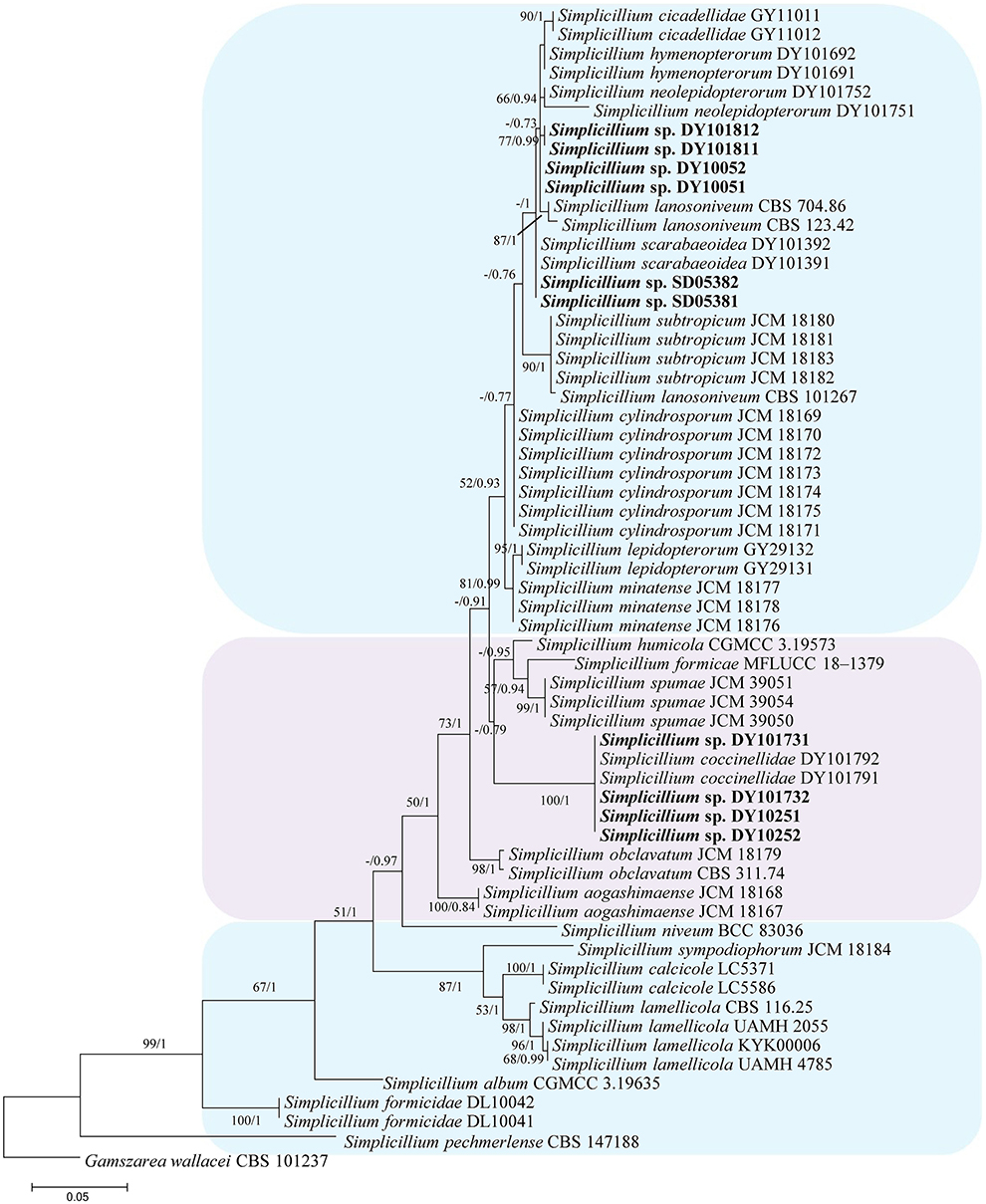
Figure 1. Phylogenetic identification of the new strains in the genus Simplicillium based on ITS sequence. Statistical support values (≥ 0.5/50%) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
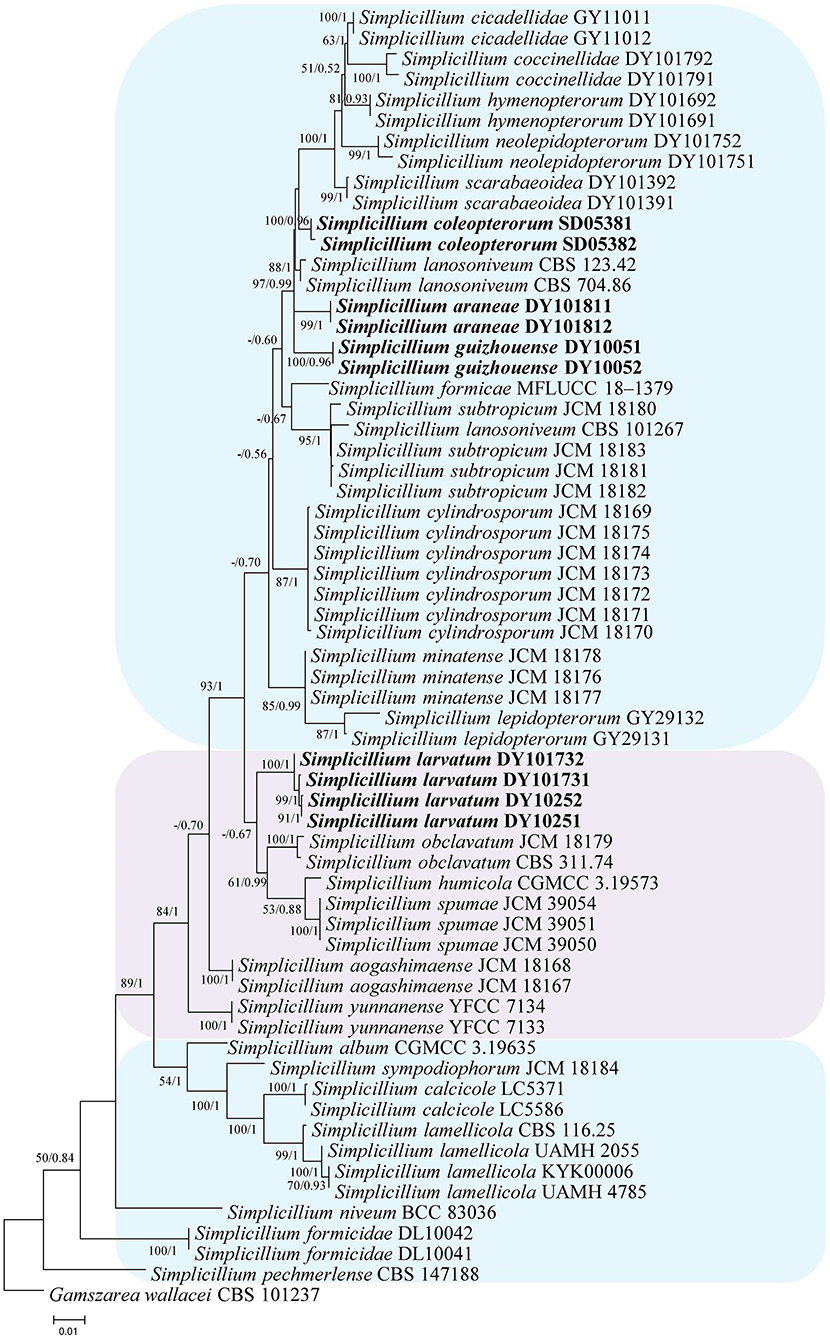
Figure 2. Phylogenetic analysis to establish the new species in the genus Simplicillium by SSU, ITS, LSU, RPB1, and TEF sequences. Statistical support values (≥ 0.5/50%) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Analysis 1: The final value of the highest scoring tree was −2,850.268342, which was obtained from an ML analysis of the ITS sequence. The parameters of the General Time Reversible (GTR) model used to analyze the dataset were estimated using the following frequencies: A = 0.230169, C = 0.278999, G = 0.261573, and T = 0.229259; substitution rates AC = 1.092678, AG = 1.861964, AT = 1.258018, CG = 0.827521, CT = 3.497952, and GT = 1.000000, as well as the gamma distribution shape parameter α = 0.302260. The selected model for BI analysis was K2P+G4 (ITS). The phylogenetic trees (Figure 1) constructed using ML and BI analyses were largely congruent and strongly supported in most branches. Strains DY10051, DY10052, DY101811, and DY101812 were grouped with Simplicillium cicadellidae W.H. Chen, C. Liu, Y.F. Han, J.D. Liang and Z.Q. Liang, S. hymenopterorum W.H. Chen et al., S. lanosoniveum, and S. neolepidopterorum W.H. Chen et al. Strains SD05381 and SD05382 were grouped with S. scarabaeoidea W.H. Chen et al. Strains DY10251, DY10252, DY101731, and DY101732 have a close relationship with S. coccinellidae W.H. Chen et al.
Analysis 2: The final value of the highest scoring tree was −17,350.543656, which was obtained from the ML analysis of the dataset (SSU+ITS+LSU+RPB1+TEF). The parameters of the GTR model used to analyze the dataset were estimated based on the following frequencies, A = 0.242448, C = 0.266436, G = 0.262106, and T = 0.229010; substitution rates AC = 1.080009, AG = 1.915942, AT = 1.147141, CG = 0.835034, CT = 5.341508, and GT = 1.000000, as well as the gamma distribution shape parameter α = 0.291522. The selected model for BI analysis was K2P+G4 (LSU) and SYM+I+G4 (SSU+ITS+RPB1+TEF). The phylogenetic trees (Figure 2) constructed using ML and BI analyses were largely congruent and strongly supported in most branches. Most genera were clustered into their independent clade. The new strains were clustered into four independent clades. Simplicillium larvatum (DY101731, DY101732, DY10251, and DY10252) had a close relationship with S. obclavatum, S. humicola Z.F. Zhang and L. Cai, and S. spumae N. Kondo, H. Iwasaki and Nonaka. S. araneae (DY101811 and DY101812), S. guizhouense (DY1005 and DY10052), and S. coleopterorum (SD05381 and SD05382) had a close relationship with S. lanosoniveum.
The topological structure of the network is consistent with that of the phylogenetic tree (Figure 1) and could be used for species relationship analysis (Huson and Bryant, 2006). However, a reticular structure was formed in the phylogenetic network by the split of information conflict or fuzzy signals. Three groups were present in the phylogenetic network (Figure 3).
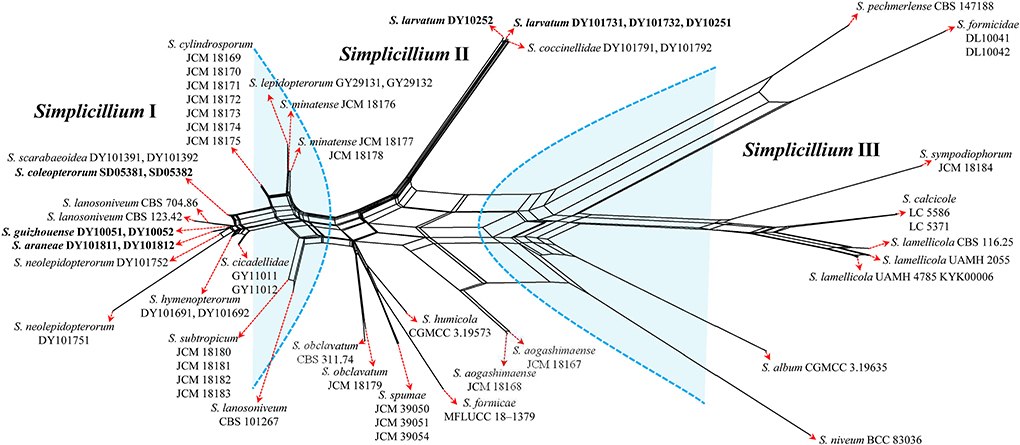
Figure 3. Reconstruction of the neighbor-net network based on ITS sequences from taxa in Figure 1.
Group I: New strains DY10051, DY10052, DY101811, DY101812, SD05381, and SD05382 grouped with Simplicillium cicadellidae (GY11011 and GY11012), S. cylindrosporum Nonaka, Kaifuchi and Masuma (JCM 18169, JCM 18170, JCM 18171, JCM 18172, JCM 18173, JCM 18174, and JCM 18175), S. hymenopterorum (DY101691 and DY101692), S. lanosoniveum (CBS 101267, CBS 123.42, and CBS 704.86), S. lepidopterorum W.H. Chen et al. (GY29131 and GY29132), S. minatense Nonaka, Kaifuchi and Masuma (JCM 18176, JCM 18177, and JCM 18178), S. neolepidopterorum (DY101751 and DY101752), and S. scarabaeoidea (DY101391 and DY101392).
Group II: S. aogashimaense Nonaka, Kaifuchi and Masuma (JCM 18167 and JCM 18168) grouped with S. formicae D.P. Wei and K.D. Hyde (MFLUCC 18–1379), S. humicola (CGMCC 3.19573), S. obclavatum (CBS 311.74 and JCM 18179), and S. spumae (JCM 39050, JCM 39051 and JCM 39054). The new strains DY10251, DY10252, DY101731 and DY101732 are clustered with S. coccinellidae (DY101791 and DY101792) into an independent subgroup.
Group III: S. album Z.F. Zhang and L. Cai (CGMCC 3.19635) grouped with S. calcicole Z.F. Zhang, F. Liu and L. Cai (LC5371 and LC5586), S. formicidae W.H. Chen et al. (DL10041 and DL10042), S. lamellicola (CBS 116.25, KYK00006, UAMH 2055 and UAMH 4785), S. niveum Mongkols., Noisrip. and Luangsa-ard (BCC 83036), S. pechmerlense J. Leplat (CBS 147188), and S. sympodiophorum Nonaka, Kaifuchi and Masuma (JCM 18184).
Taxonomy
Simplicillium araneae W.H. Chen, Y.F. Han, J.D. Liang and Z.Q. Liang, sp. nov.
MycoBank: 844146.
Type: CHINA, Guizhou, Qiannan Buyi, and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On a dead spider (Araneae), 1 October 2019, Wanhao Chen, GZAC DY10181 (holotype), ex-type living cultures, DY101811.
Description: The colonies showed moderate growth on PDA, reaching a diameter of 31–33 mm in 14 days at 25°C, were convex, with white velutinate aerial mycelium on the front and an yellowish to brown mycelium on the reverse, especially in the middle and entire margins, and soluble pigment was not produced. Vegetative hyphae were branched, hyaline, smooth-walled, septate, and 1.2–1.8 μm wide. The phialides produced on the aerial hyphae were always solitary, aseptate, hyaline, smooth-walled, relatively slender, tapering toward the tip, and 32.9–47.1 × 1.2–2.4 μm in size. Conidia hyaline was ellipsoidal to globose, aseptate, smooth-walled, 1-celled, and 1.8–2.9 × 1.2–1.8 μm in size. Octahedral crystals were absent, and a sexual state was not observed.
Etymology. Referring to the ability to colonize spiders.
Additional strain examined. China, Guizhou, Qiannan Buyi, and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On a dead spider (Araneae), 1 October 2019, Wanhao Chen, DY101812.
Notes: Simplicillium araneae was identified as belonging to Simplicillium because of its solitary phialides (Figure 4) and the analysis ITS sequence (Figure 1). Compared to the typical characteristics of 23 species, S. araneae is morphologically similar to S. formicae, S. hymenopterorum, and S. neolepidopterorum based on the absence of a slime head and octahedral crystals. However, based on the combined dataset of SSU, ITS, LSU, RPB1, and TEF sequences (Figure 2), S. araneae clustered into an independent clade and was distinguished from other Simplicillium species.
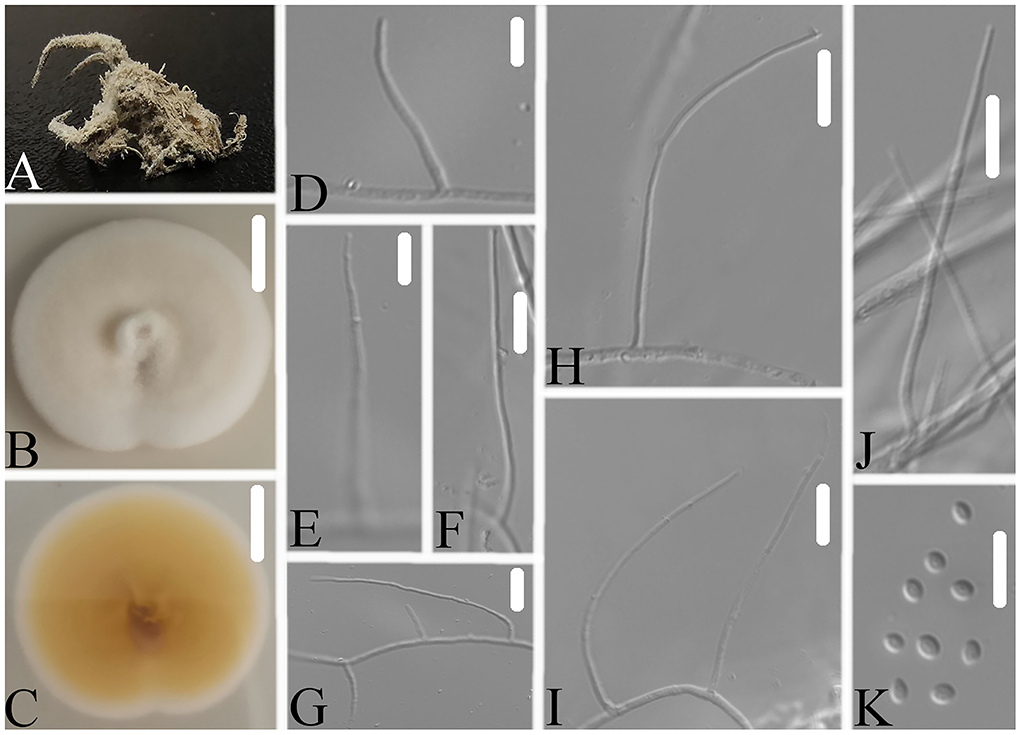
Figure 4. Simplicillium araneae (A) Infected spider (Araneae); (B,C) PDA-containing culture plate showing the front (B) and reverse (C) sides of the colony; (D–K) solitary phialides and conidia. Scale bars: 10 mm (B,C) and 10 μm (D–K).
Simplicillium coleopterorum W.H. Chen, Y.F. Han, J.D. Liang, and Z.Q. Liang, sp. nov.
MycoBank: 844147.
Type: CHINA, Guizhou, Qiannan Buyi, and Miao Autonomous Prefecture, Sandu County (25°57'22.21” N, 107°57'54.69” E). On a beetle (Coleoptera), 1 May 2019, Wanhao Chen, GZAC SD0538 (holotype), ex-type living cultures, SD05381.
Description: The colonies showed moderate growth on PDA, reaching a diameter of 49–50 mm in 14 days at 25°C, and convex, with white velutinate aerial mycelium on the front and pale brown to brown mycelium on the reverse, especially in the middle and entire margins, and soluble pigment was not produced. The vegetative hyphae were branched, hyaline, smooth-walled, septate, and 1.0–1.6 μm wide. The phialides produced on the aerial hyphae were always solitary, aseptate, hyaline, smooth-walled, relatively slender, tapering toward the tip, and 34.5–64.1 × 0.7–1.2 μm in size. Conidia were observed as small sub-globose slimy heads at the apex of the phialides, hyaline, ellipsoidal to globose in shape, aseptate, smooth-walled and 1-celled, and 2.1–3.3 × 1.5–1.9 μm in size. Octahedral crystals were absent, and a sexual state was not observed.
Etymology: Referring to an insect host in order Coleoptera.
Additional strain examined: China, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Sandu County (25°57'22.21” N, 107°57'54.69” E). On a beetle (Coleoptera), 1 May 2019, Wanhao Chen, SD05382.
Notes: Simplicillium coleopterorum was identified as belonging to Simplicillium because of its solitary phialides, conidia adhering in subglobose slimy heads, and the absence of octahedral crystals (Figure 5), supported by phylogenetic analysis of ITS sequence (Figure 1). Compared with the typical characteristics of 23 species, S. coleopterorum was morphologically similar to S. cicadellidae, S. coccinellidae, S. formicidae, S. lepidopterorum, S. niveum, S. scarabaeoidea, and S. yunnanense (Figure 6). However, based on the combined dataset of SSU, ITS, LSU, RPB1, and TEF sequences (Figure 2), S. coleopterorum was clustered into an independent clade and distinguished from S. cicadellidae, S. coccinellidae, S. formicidae, S. lepidopterorum, S. niveum, S. scarabaeoidea, and S. yunnanense.

Figure 5. Simplicillium coleopterorum (A) Infected insect (Coleoptera); (B,C) PDA-containing culture plate showing the front (B) and reverse (C) sides of the colony; (D–K) solitary phialides and conidia. Scale bars: 10 mm (B,C) and 10 μm (D–K).
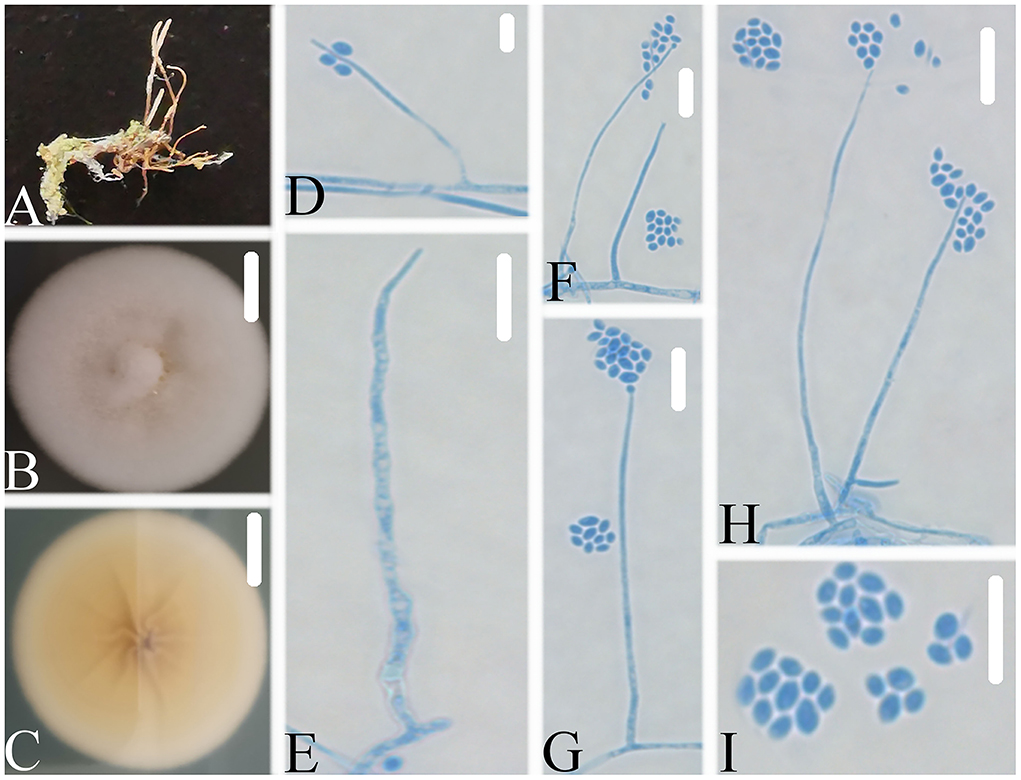
Figure 6. Simplicillium guizhouense (A) Infected ant (Formicidae); (B,C) PDA-containing culture plate showing the front (B) and reverse (C) sides of the colony; (D–I) solitary phialides and conidia. Scale bars: 10 mm (B,C) and 10 μm (D–I).
Simplicillium guizhouense W.H. Chen, Y.F. Han, J.D. Liang and Z.Q. Liang sp. nov.
MycoBank: 844148.
Type: CHINA, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On an ant (Formicidae), 1 October 2019, Wanhao Chen, GZAC DY1005 (holotype), ex-type living cultures, DY10051.
Description: The colonies showed moderate growth on PDA, reaching a diameter of 35–36 mm in 14 days at 25°C, and were convex, with white velutinate aerial mycelium on the front and yellowish to pale yellowish mycelium on the reverse, especially in the middle and entire margin, and soluble pigment was not produced. The vegetative hyphae were branched, hyaline, smooth-walled, septate, and 1.4–1.5 μm wide. The phialides produced on the aerial hyphae were always solitary, aseptate, hyaline, smooth-walled, relatively slender, and tapering toward the tip, and 21.1–52.2 × 1.0–1.8 μm in size. Conidia were observed as small globose slimy heads at the apex of the phialides, hyaline, ellipsoidal in shape, aseptate, smooth-walled and 1-celled, and 2.4–2.9 × 1.6–1.8 μm in size. Octahedral crystals were absent, and a sexual state was not observed.
Etymology: Referring to the place where the fungus was collected.
Additional strain examined: China, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On an ant (Formicidae), 1 October 2019, Wanhao Chen, DY10052.
Notes: Simplicillium guizhouense is morphologically similar to S. cicadellidae, S. coccinellidae, S. formicidae, S. lepidopterorum, S. niveum, S. scarabaeoidea, and S. yunnanense (Figure 6). Based on the combined dataset of SSU, ITS, LSU, RPB1, and TEF sequences (Figure 2), S. guizhouense clustered into an independent clade, and was distinguished from S. cicadellidae, S. coccinellidae, S. formicidae, S. lepidopterorum, S. niveum, S. scarabaeoidea, and S. yunnanense.
Simplicillium larvatum W.H. Chen, Y.F. Han, J.D. Liang and Z.Q. Liang, sp. nov.
MycoBank: 844149.
Type: CHINA, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On a larva (Lepidoptera), 1 October 2019, Wanhao Chen, GZAC DY10173 (holotype), ex-type living cultures, DY101731.
Description: The colonies showed moderate growth on PDA, reaching a diameter of 40–43 mm in 14 days at 25°C, and were convex, with white velutinate aerial mycelium on the front and yellowish mycelium on the reverse, especially in the middle and entire margin, and soluble pigment was not produced. The vegetative hyphae were branched, hyaline, smooth-walled, septate, and 0.9–1.6 μm wide. The phialides produced on the aerial hyphae were always solitary, aseptate, hyaline, smooth-walled, relatively slender, tapering toward the tip, and 16.4–28.7 × 1.2–1.7 μm in size. Conidia were observed as small sub-globose slimy heads at the apex of the phialides, hyaline, ellipsoidal to long ellipsoidal in shape, aseptate, smooth-walled and 1-celled, and 1.8–3.3 × 1.6–2.0 μm in size. Octahedral crystals were absent, and a sexual state was not observed.
Etymology: Referring to its insect host, a larva.
Additional strain and specimen examined. China, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21'27.96”N, 107°22'48.22”E). On a larva (Lepidoptera), 1 October 2019, Wanhao Chen, DY101732; on a pupa (Lepidoptera), 1 October 2019, Wanhao Chen, GZAC DY1025, living cultures, DY10251, DY10252.
Notes: Simplicillium larvatum is morphologically similar to S. cicadellidae, S. coccinellidae, S. formicidae, S. lepidopterorum, S. niveum, S. scarabaeoidea, and S. yunnanense (Figure 7). However, based on the combined dataset of SSU, ITS, LSU, RPB1, and TEF sequences (Figure 2), S. larvatum was clustered into an independent clade and phylogenetically close to S. humicola, S. obclavatum, and S. spumae. However, S. larvatum was morphologically distinguished from S. humicola, which has bigger conidia (3.0–5.0 × 1.5–3.0 μm) with octahedral crystals present. S. larvatum was morphologically distinguished from S. obclavatum, which has longer phialide (30–52 × 0.8–2.0 μm) with octahedral crystals present. S. larvatum was morphologically distinguished from S. spumae, which has subglobose or oval to ellipsoidal and octahedral crystals present.
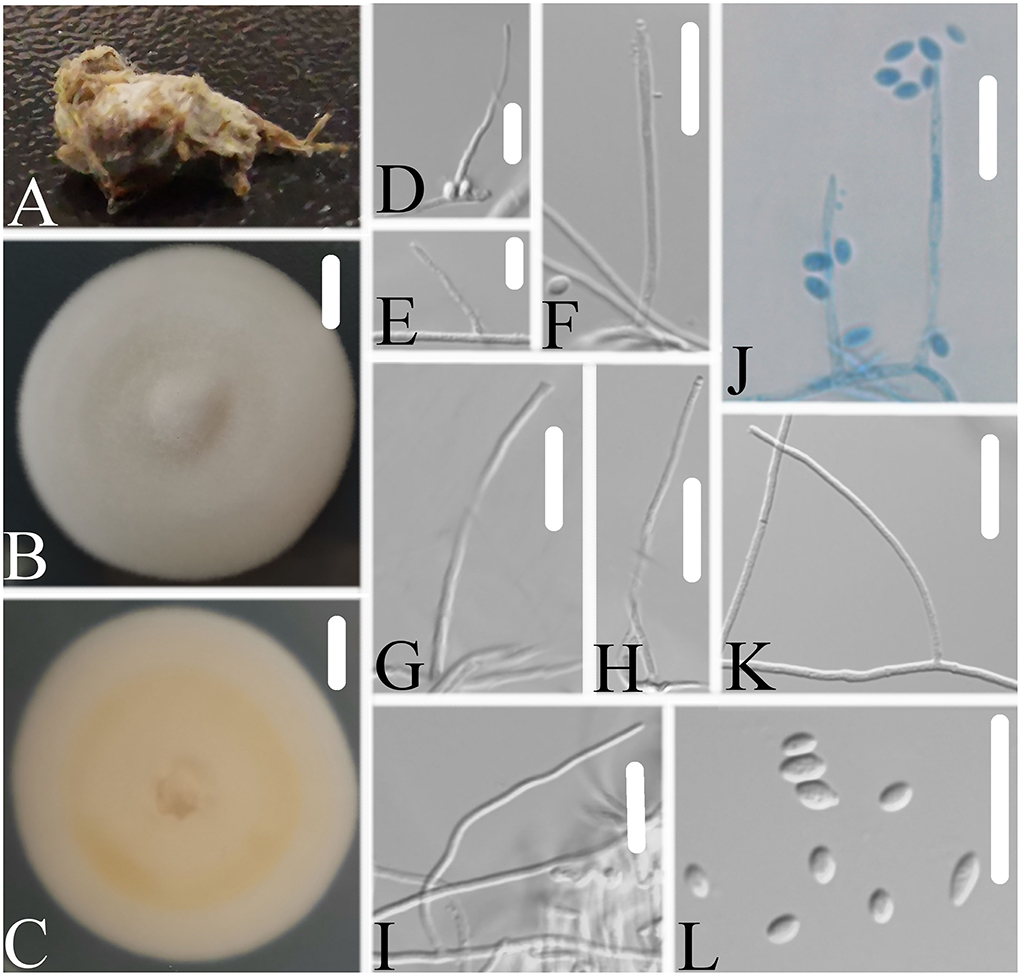
Figure 7. Simplicillium larvatum (A) Infected larva (Lepidoptera); (B,C) PDA-containing culture plate showing the front (B) and reverse (C) sides of the colony; (D–L) solitary phialides and conidia. Scale bars: 10 mm (B,C) and 10 μm (D–L).
Discussion
Zare and Gams (2001) noted that Simplicillium species are widely distributed and commonly found on various substrates or as fungicolous fungi. S. album, S. calcicola, S. cylindrosporum, S. humicola, S. minatense, S. obclavatum, S. pechmerlense, S. subtropicum, and S. sympodiophorum were isolated from soil, marine water, rock, decaying wood, and air (Zare and Gams, 2001; Liu and Cai, 2012; Nonaka et al., 2013; Liang et al., 2017; Zhang et al., 2020b; Leplat et al., 2021). S. aogashimaense was isolated from the soil and has also been reported as a symbiotic fungus (Nonaka et al., 2013; Shentu et al., 2020). S. lanosoniveum was reported as both an endophytic and an hyperparasitic fungus (Baiswar et al., 2014; Wei et al., 2019). S. formicae, S. lamellicola, S. niveum, and S. yunnanense were reported as hyperparasitic fungi (Shin et al., 2017; Wei et al., 2019; Wang et al., 2020; Crous et al., 2021). S. cicadellidae, S. coccinellidae, S. formicidae, S. hymenopterorum, S. lepidopterorum, S. neolepidopterorum, and S. scarabaeoidea were reported to be associated with different insects (Chen et al., 2019, 2021). In the present study, four new species associated with spiders and other insect substrates have been reported. Thus, Simplicillium species completely fit with the nutritional model of Hypocreales fungi and may be used as a model to study their evolutionary relationship.
In the present study, the phylogenetic network was reconstructed to explore the evolutionary relationship, consistent with the phylogenetic tree of the ITS sequence and the combined datasets (SSU, ITS, LSU, RPB1, TEF). According to the phylogenetic network (Figure 3), Simplicillium guizhouense (DY10051 and DY10052) may share a common ancestor with S. araneae (DY101811 and DY101812), S. lanosoniveum (CBS 123.42 and CBS 704.86), and S. neolepidopterorum (DY101751 and DY101752). S. lepidopterorum (GY29131 and GY29132) may share a common ancestor with S. minatense (JCM 18176, JCM 18177, and JCM 18178). S. obclavatum (CBS 311.74 and JCM 18179) may share a common ancestor with S. formicae (MFLUCC 18–1379), S. humicola (CGMCC 3.19573), and S. spumae (JCM 39050, JCM 39051 and JCM 39054). S. calcicole (LC5371 and LC5586) may share a common ancestor with S. lamellicola (CBS 116.25, UAMH 2055, KYK00006, UAMH 4785).
Host shift is usually described as an evolutional process between fungi and their hosts and is often determined by nutrient requirements (Vega et al., 2009). The nutritional model of Hypocreales fungi goes from plants (including living plants and plant residues) to animals (especially insects) and finally to fungi (Spatafora et al., 2007). The results of the phylogenetic network suggest that S. araneae, S. lanosoniveum, and S. neolepidopterorum may have originally come from insects and then jumped to a spider host, plant and fungi substrate, or another insect host, respectively. S. lepidopterorum may have originally come from the soil and then jumped to an insect host. S. formicae and S. spumae may have originally come from air or soil and then jumped as hyperparasitic fungi or water substrates. S. lamellicola may have originally come from rock substrate and then jumped as hyperparasitic fungus. These results suggest that host jump may be common in Simplicillium species.
S. coleopterorum and S. larvatum could not split from S. scarabaeoidea and S. coccinellidae in the phylogenetic network as only the ITS sequence was analyzed. However, they could be phylogenetically distinguished by a multi-locus phylogenetic analysis. Therefore, more sequence information should be added to the phylogenetic network in order to analyze their evolutionary relationship. Moreover, S. lanosoniveum was transferred to the genus Simplicillium by Zare and Gams (2001) with the strain CBS 123.42. In the present study, three strains of S. lanosoniveum (CBS 101267, CBS 123.42, and CBS 704.86) were tested. Strains CBS 123.42 and CBS 704.86 were clustered into a subclade. However, strain CBS 101267 was clustered with four strains of S. subtropicum (JCM 18180, JCM 18181, JCM18182, and JCM 18183). The pairwise dissimilarity of ITS sequences shows only a 4 bp difference, with 552 bp between strains CBS 101267 and JCM18180. This result supports strain CBS 101267 being identified as S. subtropicum.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
WC isolated the fungi, built up the phylogenetic tree, and wrote the manuscript. YH identified the fungal isolates, revised the manuscript, and provided partial funding. JL, XR, and JZ revised the manuscript and provided partial funding. ZL identified the fungal isolates and revised the manuscript. All authors discussed the results.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31860002), the High-level Innovative Talents Training Object in Guizhou Province (No. Qiankehepingtairencai [2020]6005), the Science and Technology Foundation of Guizhou Province (No. Qiankehejichu [2020]1Y060), the Program of Innovative Scientific and Technological Talent Team of Guizhou Province (2020-5010), and the Construction Program of Guizhou Engineering Research Center (Qian Fa Gai Gao Ji 2020-896).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.950773/full#supplementary-material
References
Baiswar, P., Ngachan, S. V., Rymbai, H., and Chandra, S. (2014). Simplicillium lanosoniveum, a hyperparasite on Aecidium elaeagni-latifoliae in India. Australas Plant Dis. Notes 9, 1–5. doi: 10.1007/s13314-014-0144-z
Bandelt, H. J., and Dress, A. W. (1992). Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1, 242–252. doi: 10.1016/1055-7903(92)90021-8
Bryant, D., and Moulton, V. (2004). Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265. doi: 10.1093/molbev/msh018
Castlebury, L. A., Rossman, A. Y., Sung, G. H., Hyten, A. S., and Spatafora, J. W. (2004). Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872. doi: 10.1017/S0953756204000607
Chen, W. H., Han, Y. F., Liang, J. D., and Liang, Z. Q. (2021). Taxonomic and phylogenetic characterizations reveal four new species of Simplicillium (Cordycipitaceae, Hypocreales) from Guizhou, China. Sci. Rep 11, 15300. doi: 10.1038/s41598-021-94893-z
Chen, W. H., Liu, C., Han, Y. F., Liang, J. D., Tian, W. Y., et al. (2019). Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from Southwest China. MycoKeys 58, 83–102. doi: 10.3897/mycokeys.58.37176
Cheng, C. H., and Huang, Y. (2008). Construction and application of phylogenetic network. Entomotaxonomia 03, 215–221.
Crous, P. W., Cowan, D. A., Maggs-Kölling, G., Yilmaz, N., Thangavel, R., et al. (2021). Fungal planet description sheets: 1182-1283. Persoonia 46, 313–528. doi: 10.3767/persoonia.2021.46.11
Crous, P. W., Luangsa-ard, J. J., Wingfield, M. J., Carnegie, A. J., Hernández-Restrepo, M., et al. (2018). Fungal Planet description sheets: 785–867. Persoonia 41, 238. doi: 10.3767/persoonia.2018.41.12
Drummond, A., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Gams, W. (2017). An annotated checklist of epithets published in Verticillium and Acrostalagmus, some similar genera, and teleomorphs associated with verticillium-like anamorphs. Nova Hedwigia 104, 381–450. doi: 10.1127/nova_hedwigia/2016/0371
Gomes, A. A., Pinho, D. B., Cardeal, Z. L., Menezes, H. C., De Queiroz, M. V., et al. (2018). Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa 333, 188–198. doi: 10.11646/phytotaxa.333.2.2
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khonsanit, A., Luangsa-ard, J. J., Thanakitpipattana, D., Noisripoom, W., Chaitika, T., et al. (2020). Cryptic diversity of the genus Beauveria with a new species from Thailand. Mycol. Prog. 19, 291–315. doi: 10.1007/s11557-020-01557-9
Kondo, N., Iwasaki, H., Tokiwa, T., Omura, S., and Nonaka, K. (2020). Simplicillium spumae (Cordycipitaceae, Hypocreales), a new hyphomycetes from aquarium foam in Japan. Mycoscience 61, 116–121. doi: 10.1016/j.myc.2020.02.002
Leplat, J., Francois, A., and Bousta, F. (2021). Simplicillium pech-merlensis, a new fungal species isolated from the air of the Pech-Merle show cave. Phytotaxa 521, 80–94. doi: 10.11646/phytotaxa.521.2.2
Liang, J. D., Han, Y. F., Zhang, J. W., Du, W., Liang, Z. Q., et al. (2011). Optimal culture conditions for keratinase production by a novel thermophilic Myceliophthora thermophila strain GZUIFR-H49-1. J. Appl. Microbiol. 110, 871–880. doi: 10.1111/j.1365-2672.2011.04949.x
Liang, X., Qi, S. H., Nong, X. H., and Huang, Z. H. (2017). Antifungal and antiviral cyclic peptides from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. J. Agric. Food Chem. 65, 5114–5121. doi: 10.1021/acs.jafc.7b01238
Liu, F., and Cai, L. (2012). Morphological and molecular characterization of a novel species of Simplicillium from China. Cryptogamie, Mycologie 33, 137–145. doi: 10.7872/crym.v33.iss2.2012.137
Morozov, A. A., Galachyants, Y. P., and Likhoshway, Y. V. (2013). Inferring phylogenetic networks from gene order data. Biomed. Res. Int. 2013, 1–7. doi: 10.1155/2013/503193
Morrison, D. A. (2005). Networks in phylogenetic analysis: new tools for population biology. Int. J. Parasitol. 35, 567–582. doi: 10.1016/j.ijpara.2005.02.007
Nonaka, K., Kaifuchi, S., Omura, S., and Masuma, R. (2013). Five new Simplicillium species (Cordycipitaceae) from soils in Tokyo, Japan. Mycoscience 54, 42–53. doi: 10.1016/j.myc.2012.07.002
Okane, I., Nonaka, K., Kurihara, Y., Abe, J. P., and Yamaoka, Y. (2020). A new species of Leptobacillim, L. symbioticum, isolated from mites and sori of soybean rust. Mycoscience 61, 165–171. doi: 10.1016/j.myc.2020.04.006
Rakotonirainy, M. S., Cariou, M. L., Brygoo, Y., and Riba, G. (1994). Phylogenetic relationships within the genus Metarhizium based on 28S rRNA sequences and isozyme comparison. Mycol. Res. 98, 225–230. doi: 10.1016/S0953-7562(09)80190-1
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shentu, X., Xiao, Y., Song, Y., Cao, Z., Fan, J., et al. (2020). Comparative analysis of the diversity of the microbial communities between non-fertilized and fertilized eggs of brown planthopper, Nilaparvata lugens Stål. Insects 11, 49. doi: 10.3390/insects11010049
Shin, T. S., Yu, N. H., Lee, J., Choi, G. J., Kim, J. C., et al. (2017). Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and ginseng. Plant Pathol. J. 33, 337. doi: 10.5423/PPJ.FT.04.2017.0087
Silvestro, D., and Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers Evol. 12, 335–337. doi: 10.1007/s13127-011-0056-0
Spatafora, J. W., Sung, G. H., Sung, J. M., Hywel-Jones, N. L., and White, J. F. (2007). Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 16, 1701–1711. doi: 10.1111/j.1365-294X.2007.03225.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vega, F. E., Goettel, M. S., Blackwell, M., Chandler, D., Jackson, M. A., et al. (2009). Fungal entomopathogens: new insights on their ecology. Fungal. Ecol. 2, 149–159. doi: 10.1016/j.funeco.2009.05.001
Wang, Y. B., Wang, Y., Fan, Q., Duan, D. E., Zhang, G. D., et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 103, 1–46. doi: 10.1007/s13225-020-00457-3
Wei, D. P., Wanasinghe, D. N., Hyde, K. D., Mortimer, P. E., Xu, J., et al. (2019). The genus Simplicillium. MycoKeys 60, 69–92. doi: 10.3897/mycokeys.60.38040
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J. (Eds.) PCR protocols: a guide to methods and applications. New York, NY: Academic Press. p. 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Zare, R., and Gams, W. (2001). A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia. 73, 1–50. doi: 10.1127/nova.hedwigia/73/2001/1
Zare, R., and Gams, W. (2008). A revision of the Verticillium fungicola species complex and its affinity with the genus Lecanicillium. Mycol. Res. 112, 811–824. doi: 10.1016/j.mycres.2008.01.019
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., et al. (2020a). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, Z. F., Liu, F., Zhou, X., Liu, X. Z., Liu, S. J., et al. (2017). Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39, 1–31. doi: 10.3767/persoonia.2017.39.01
Zhang, Z. F., Zhou, S. Y., Eurwilaichitr, L., Ingsriswang, S., Raza, M., et al. (2020b). Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers 106, 29–136. doi: 10.1007/s13225-020-00453-7
Keywords: spider, insect, multigene phylogeny, morphological characterization, phylogenetic relationship
Citation: Chen W, Liang J, Ren X, Zhao J, Han Y and Liang Z (2022) Multigene phylogeny, phylogenetic network, and morphological characterizations reveal four new arthropod-associated Simplicillium species and their evolutional relationship. Front. Microbiol. 13:950773. doi: 10.3389/fmicb.2022.950773
Received: 23 May 2022; Accepted: 29 July 2022;
Published: 04 October 2022.
Edited by:
F. L. Consoli, University of São Paulo, BrazilReviewed by:
Josepa Gené, University of Rovira i Virgili, SpainNicolau Sbaraini, Federal University of Rio Grande do Sul, Brazil
Copyright © 2022 Chen, Liang, Ren, Zhao, Han and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfeng Han, swallow1128@126.com
 Wanhao Chen
Wanhao Chen Jiandong Liang1
Jiandong Liang1 Zongqi Liang
Zongqi Liang