- 1Yunnan Herbal Laboratory, College of Ecology and Environmental Sciences, Yunnan University, Kunming, China
- 2The International Joint Research Center for Sustainable Utilization of Cordyceps Bioresources in China and Southeast Asia, Yunnan University, Kunming, China
- 3Faculty of Agricultural Production, Maejo University, Chiang Mai, Thailand
- 4Institute of Regional Research and Development, Ministry of Science and Technology, Hanoi, Vietnam
Introduction: Clonostachys, a genus with rich morphological and ecological diversity in Bionectriaceae, has a wide distribution among diverse habitats.
Methods and Results: In the present study, a phylogenetic framework is reconstructed for the family Bionectriaceae focusing on Clonostachys through increased taxon-sampling using the nrLSU sequence. Through surveying Clonostachys in China, Vietnam, and Thailand over the past 3 years, seven Clonostachys spp. were found and identified. Two new species, C. chuyangsinensis and C. kunmingensis, are described and illustrated based on morphological characteristics and molecular data. The phylogenetic positions of the seven species were evaluated based on four genomic loci (ITS, nrLSU, TUB2, and TEF1).
Discussion: Moreover, the genetic divergence comparisons of Clonostachys species for three markers (ITS, TUB2, and TEF1) are also provided. The results indicated that the TEF1 sequence data provided the best resolution for distinguishing species of Clonostachys, followed by sequence data for the TUB2 and ITS regions.
Introduction
The asexual morph-typified genus Clonostachys was established by Corda (1839) on the basis of the type species, C. araucaria, which possessed penicillate conidiophores and imbricate conidia held in columns. This species is now considered a synonym of C. rosea (Link) Schroers et al. (basionym Penicillium roseum Link) (Schroers et al., 1999). Clonostachys (Bionectriaceae, Hypocreales) is characterized by penicillate, sporodochial, or dimorphic conidiophores and phialidic conidiogenous cells producing hyaline conidia (Schroers, 2001). Teleomorph, is originially described Bionectria (Spegazzini, 1919), and characterized by ascomata typically seated on a pseudoparenchymatous stroma or arising directly on the substrate, being white, pale yellow, or orange to dark brownish-orange, not changing color in 3% KOH or lactic acid, not collapsing or laterally pinched when dry; warted or smooth; an ascomatal wall composed of 1–3 regions with the outer region composed of subglobose to globose, thick-walled cells; ascospores smooth; spinulose, striate or warted (Schroers, 2001; Lechat and Fournier, 2018). Based on the monograph of Bionectria and Clonostachys by Schroers (2001), this connection was confirmed by DNA sequences. Because Clonostachys was described earlier than Bionectria, Rossman et al. (2013) recommended Clonostachys as the name of this genus.
It is generally agreed that distinguishing individual species of Clonostachys using only morphological characteristics can be difficult (Schroers et al., 1999; Abreu et al., 2014). The members of the genus Clonostachys were accommodated in Acrostalagmus, Clonostachyopsis, Dendrodochium, Gliocladium, Gliocladochium, Myrothecium, Sesquicillium, Spicaria, Verticilliodochium, or Verticillium (Schroers, 2001). It is the huge diversification of morphs of closely related Clonostachys species, what did not allow recognition that they all may belong to a single genus, Clonostachys. Given the problems with species delimitation in Clonostachys using morphology, molecular data are essential to establish robust species boundaries. The first molecular study of Clonostachys/Bionectria was carried out by Rossman et al. (2001) using large subunit rDNA sequences. The results showed that the genus represents a well-resolved monophyletic lineage. Subsequently, DNA sequences of the internal transcribed spacer regions of the rDNA (ITS rDNA) and a portion of the β-tubulin (TUB2) gene were widely used to resolve taxonomic questions for Clonostachys/Bionectria (Schroers, 2001; Hirooka and Kobayashi, 2007; Luo and Zhuang, 2010; Chen et al., 2016; Prasher and Chauhan, 2017). Regrettably, not all recognized species inside this group formed well-supported clades in these two-gene phylogenies (Moreira et al., 2016). Other DNA sequences recently employed to improve the resolution of phylogenetic trees for the species of Clonostachys/Bionectria include ATP citrate lyase (ACL1), TUB2, the large subunit of RNA polymerase II (RPB1), and the translation elongation factor 1-α (TEF1) gene regions (Moreira et al., 2016). However, sequence data of the above-mentioned four protein-encoding gene regions in GenBank1 are incomplete for the group.
There is no doubt that Clonostachys belongs to the family Bionectriaceae, but its taxonomic position in relation to other genera is debated within Bionectriaceae (Rossman et al., 2001; Hyde et al., 2020; Schoch et al., 2020). In more recent studies, Clonostachys was suggested as a close relative of the genus Stephanonectria that was confirmed as a member of Bionectriaceae (Hyde et al., 2020). However, Schoch et al. (2020) reported that Stephanonectria was a genus of ascomycetes in the family Nectriaceae (2accessed on 1 July 2022). Rossman et al. (2001) found that the genera Emericellopsis and Stanjemonium belonged to Bionectriaceae in spite of the distant relation to Clonostachys, whereas Schoch et al. (2020) placed their taxonomic positions in the Hypocreales genera, incertae sedis genera (see Footnote 2 accessed on 1 July 2022). Therefore, it is imperative to reconstruct the phylogenetic framework for the Bionectriaceae focusing on Clonostachys through increased taxon sampling.
In the current study, we aimed to: (1) consider the identity of previously unidentified Clonostachys isolates collected over a 3-year period from China, Vietnam, and Thailand and (2) re-evaluate the taxonomic stability of Clonostachys among related genera within Bionectriaceae and phylogenetic relationships between Clonostachys species.
Materials and methods
Soil and specimen collection and fungus isolation
Soil samples and fungus-infected spider specimens were collected from 11 locations in 2017 and 2019, including eight different locations within Yunnan Province, China, two locations within Dak Lak Province, Vietnam, and one location in Chiang Mai, Thailand.
Clonostachys strains were isolated from the soil samples according to methods described in our previous publication (Wang et al., 2015). Briefly, 2 g of soil was added to a flask containing 20 ml sterilized water and glass beads. The soil suspension was shaken for about 10 min and then diluted 100 times. Subsequently, 200 μL of the diluted soil suspension was spread on Petri dishes with solidified onion garlic agar (OGA: 20 g of grated garlic and 20 g of onion were boiled in 1 l of distilled water for 1 h; the boiled biomass was then filtered-off, and 2% agar was added). Czapek yeast extract agar (CYA, Advanced Technology and Industrial Co., Ltd., China) and potato dextrose agar (PDA, Difco, United States) were used, and all media had 50 mg/L rose Bengal and 100 mg/L kanamycin added. Conidia developing on spider cadavers were transplanted onto plates of PDA and cultured at 25°C. Colonies of the isolated filamentous fungi appearing in the culture were transferred onto fresh PDA media. The purified fungal strain was transferred to PDA slants and cultured at 25°C until its hyphae spread across the entire slope. The emerging fungal spores were washed with sterile physiological saline and made into a spore suspension of 1 × 103 cells/mL. To obtain monospore cultures, a part of the spore suspension was placed on PDA using a sterile micropipette, and then a Petri dish was incubated at 25°C. Specimens and type material were deposited in the Yunnan Herbal Herbarium (YHH) at the Institute of Herb Biotic Resources of Yunnan University, China. Cultures were stored in the Yunnan Fungal Culture Collection (YFCC) at the Institute of Herb Biotic Resources of Yunnan University.
Morphological observations
Macroscopic characters were collected from colonies grown on PDA and corn meal agar (CMA, Shanghai yiyan bio-technology Co., Ltd., China). Cultures on PDA slants were transferred to PDA and CMA plates and incubated at 25°C for 7 days. Reverse colony pigmentation of strains grown on PDA and CMA was assessed according to Kornerup and Wanscher (1978). For morphological evaluation, microscope slides were prepared by placing mycelia from the cultures on PDA and CMA blocks (5 mm diameter) and then overlaid with a coverslip. The sizes and shapes of the microcharacteristics (e.g., ascomata, asci, ascospores, conidiogenous cells, and conidia) were determined using a light microscope (CX40, Olympus Corporation, Tokyo, Japan) and a scanning electron microscope (Quanta 200 FEG, FEI Company, Hillsboro, United States). Individual length and width measurements were taken for 30–100 replicates, including the absolute minima and maxima.
DNA extraction, polymerase chain reaction, and sequencing
Specimens and live axenic cultures were prepared for DNA extraction. Genomic DNA was extracted using the Genomic DNA Purification Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol. The primer pair ITS5 and ITS4 was used to amplify the nuclear ribosomal internal transcribed spacer region (ITS) (White et al., 1990). For amplification of the nuclear ribosomal large subunit (nrLSU) and the β-tubulin (TUB2) gene, PCR primer pairs LR5/LR0R and T1/Bt2b (Vilgalys and Hester, 1990; Rehner and Samuels, 1994; Glass and Donaldson, 1995; O’Donnell and Cigelnik, 1997) were employed. The translation elongation factor 1α (TEF1) gene was amplified using the primer pair EF1-688F/EF1-1251R (Alves et al., 2008). All of the PCR reactions were performed in a final volume of 50 μL containing 25 μL 2 × Taq PCR Master Mix (Tiangen Biotech Co., LTD, China), 0.5 μL of each primer (10 μM), 1 μL of genomic DNA, and 23 μL of RNase-Free water. PCR products were sequenced by Beijing Sinogenomax Co. Ltd., China.
Phylogenetic analyses
Phylogenetic analyses were based on the nrLSU and combined ITS+nrLSU + TUB2 + TEF1 sequences. Sequences of ITS, nrLSU, TUB2, and TEF1 were retrieved from GenBank and combined with those generated in our study. The taxonomic information and GenBank accession numbers are provided in Table 1. Sequences were aligned using Clustal X 2.0 and MEGA v6.06 software (Larkin et al., 2007; Tamura et al., 2013). After alignment, the sequences of the genes were concatenated. Conflicts among the six genes were tested using PAUP* 4.0b10 (Swofford, 2002). The results showed that the phylogenetic signals for the four loci were congruent (p = 0.03). Phylogenetic analyses were conducted using the Bayesian Inference (BI) and the Maximum Likelihood (ML) methods employing MrBayes v3.1.2 and RAxML 7.0.3 (Ronquist and Huelsenbeck, 2003; Stamatakis et al., 2008). Models of sequence evolution were estimated using jModelTest version 2.1.4 (Darriba et al., 2012). The following models were implemented in the Bayesian phylogenetic analyses: GTR + I + G for ITS and nrLSU, K80 + G for TUB, SYM + G for TEF1. The BI analysis was run on MrBayes v3.1.2 for 5 million generations. GTR + I was selected as the optimal model for ML analysis, and 1,000 rapid bootstrap replicates were performed on the dataset. Furthermore, ML analysis was applied to single-locus genealogies for ITS, nrLSU, TUB2, and TEF1.
We applied a (phylo-) genetic distance matrix calculation for the markers (ITS, TUB2, and TEF1) to assess species boundaries of 14 Clonostachys spp. (Supplementary Tables S1–S3), because their sequence data for the three loci were complete. The pairwise genetic distances of the 14 Clonostachys lineages were measured based on the Kimura two-parameter model using MEGA v6.06 software (Tamura et al., 2013).
Results
Sequencing and phylogenetic analyses
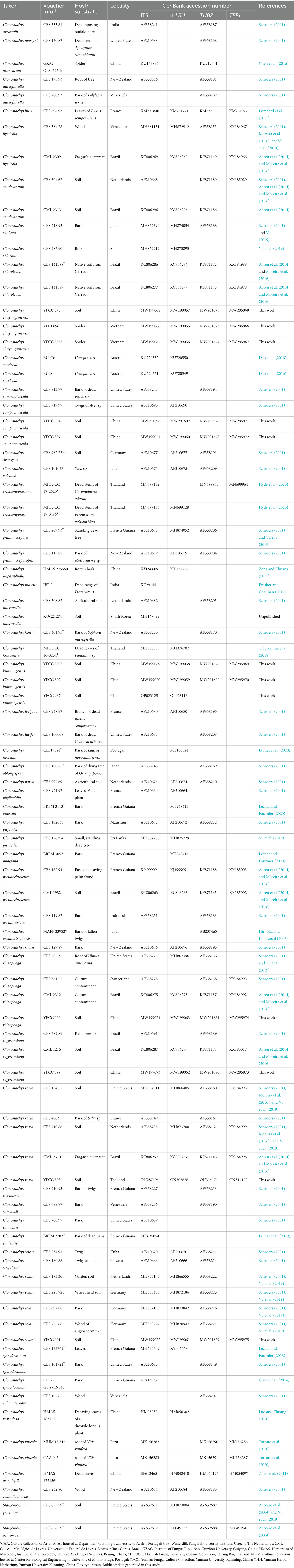
Phylogenetic analyses based on nrLSU data consisting of 107 fungal taxa confirmed the presence and positions of Clonostachys and related genera within Bionectriaceae. Eighteen well-supported clades were recognized based on both BI and ML analyses of the 107 taxa from Bionectriaceae and Flammocladiella (Flammocladiaceae, Hypocreales) that accommodate species of the genera Bryocentria, Clonostachys, Emericellopsis, Gliomastix, Heleococcum, Hydropisphaera, Ijuhya, Lasionectria, Nectriopsis, Paracylindrocarpon, Roumegueriella, Selinia, Stanjemonium, Stephanonectria, Stilbocrea, Stromatonectria, Verrucostoma, and Flammocladiella (Figure 1). The genus Clonostachys was phylogenetically clustered with Stephanonectria, and Emericellopsis had a close genetic relationship with Stanjemonium, but they were clearly distinguished from their allied genera by forming four separate clades in the family Bionectriaceae (Figure 1). The combined dataset included sequences from 86 fungal taxa (Table 1). The final dataset consisted of 2,900 bp of sequence data, including gaps (ITS, 654 bp; nrLSU, 903 bp; TUB2, 711 bp; and TEF1, 632 bp). Both BI and ML analyses produced trees with similar topologies that resolved most Clonostachys lineages in separate terminal branches (Figure 2). Phylogenetic trees inferred from analyses of combined data divided Clonostachys into six distinguished clades, designated as Astromata, Bionectria, Epiphloea, Myronectria, Uniparietina, and Zebrinella clades (Figure 2). The phylogenetic analyses suggested the existence of distinct species in the Bionectria and Epiphloea clade that we accordingly propose as new species: C. chuyangsinensis, which was found in the Epiphloea clade, and C. kunmingensis, which was found in the Bionectria clade (Figure 2).
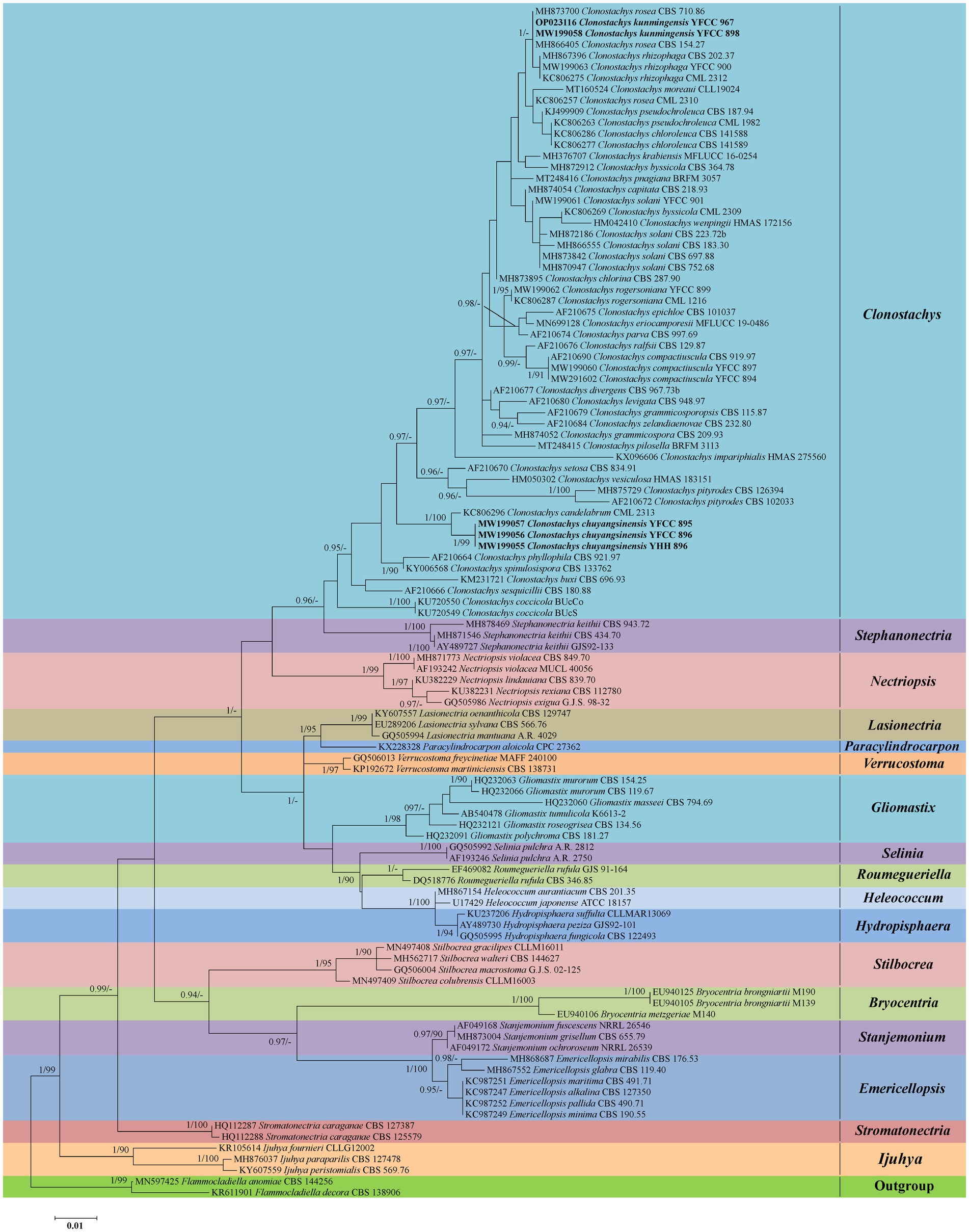
Figure 1. Phylogenetic reconstruction of Clonostachys and related genera in Bionectriaceae obtained from the nrLSU sequences based on Bayesian inference and Maximum Likelihood analyses. Statistical support values (≥0.9/90%) are shown at the nodes for BI posterior probabilities/ML bootstrap support. Materials in bold type are those analyzed in this study.
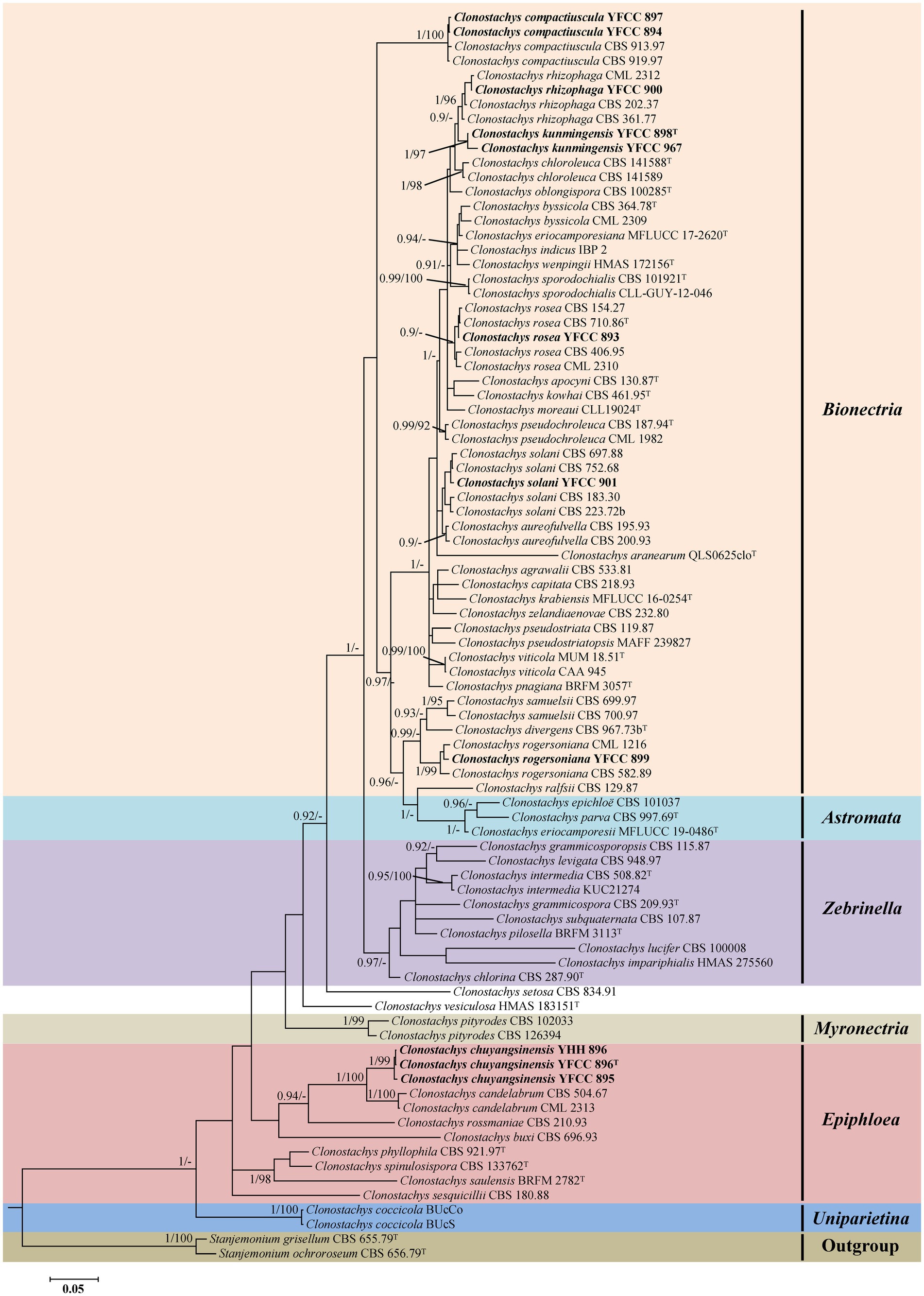
Figure 2. Phylogenetic tree of Clonostachys based on Bayesian inference and Maximum Likelihood analyses of a 4-locus (ITS, nrLSU, TUB2, and TEF1) dataset. Statistical support values (≥ 0.9/90%) are shown at the nodes for BI posterior probabilities/ML bootstrap support. Materials in bold type are those analyzed in this study. Isolates representing ex-type material are marked with “T.”
The tree topologies for the individual loci (ITS, nrLSU, TUB2, and TEF1) did not show congruence (Supplementary Figures S1–S4). However, in all analyses C. chuyangsinensis had a close genetic relationship with C. candelabrum. Clonostachys chloroleuca and C. rhizophaga were sisters to the newly discovered species C. kunmingensis, although this relationship received significant bootstrap support only from ITS and TUB2. Phylogenetic analyses based on nrLSU data revealed that C. kunmingensis was closely related to C. rosea (Figure 1; Supplementary Figure S2). And the nrLSU sequences cannot distinguish the two species. But they were regarded as different species with strong support from ITS, TUB2 and TEF1 (Supplementary Figures S1, S3, S4).
The genetic divergence comparisons showed that: (1) the minimum thresholds (p-distances) to distinguish genetic species in the Clonostachys lineages were 0.005, 0.017, and 0.026 for ITS, TUB2, and TEF1, respectively (Supplementary Tables S1–S3); (2) the TEF1 sequence data provided the best resolution distinguishing Clonostachys spp., followed by TUB2 and ITS sequences (Supplementary Tables S1–S3); and (3) the genetic distances strongly supported recognition of C. chuyangsinensis and C. kunmingensis as two new taxa (Table 2).
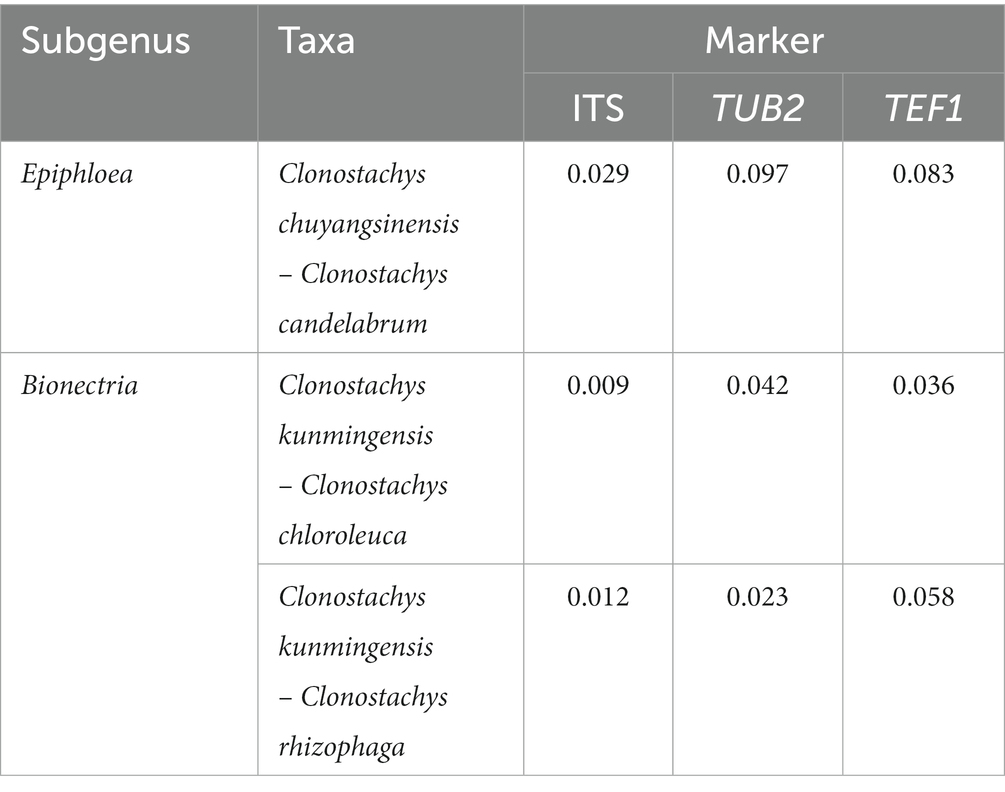
Table 2. Genetic distance (p-distances) of the two new Clonostachys species with their related species.
Taxonomy
In this study, a collection of 23 isolates of unknown identity were shown to represent five known species and two new species of Clonostachys. The phylogenetic positions of the five known species were evaluated according to phylogenetic inferences based on four loci (ITS, nrLSU, TUB2, and TEF1), including C. compactiuscula, C. rhizophaga, C. rogersoniana, and C. solani from China, and C. rosea from Thailand (see Table 1; Figure 2). The two new species, provided with the names C. chuyangsinensis from Vietnam and China and C. kunmingensis from China, were recognized based on morphological characteristics and molecular data.
Clonostachys chuyangsinensis H. Yu & Y. Wang, sp. nov. Figure 3.
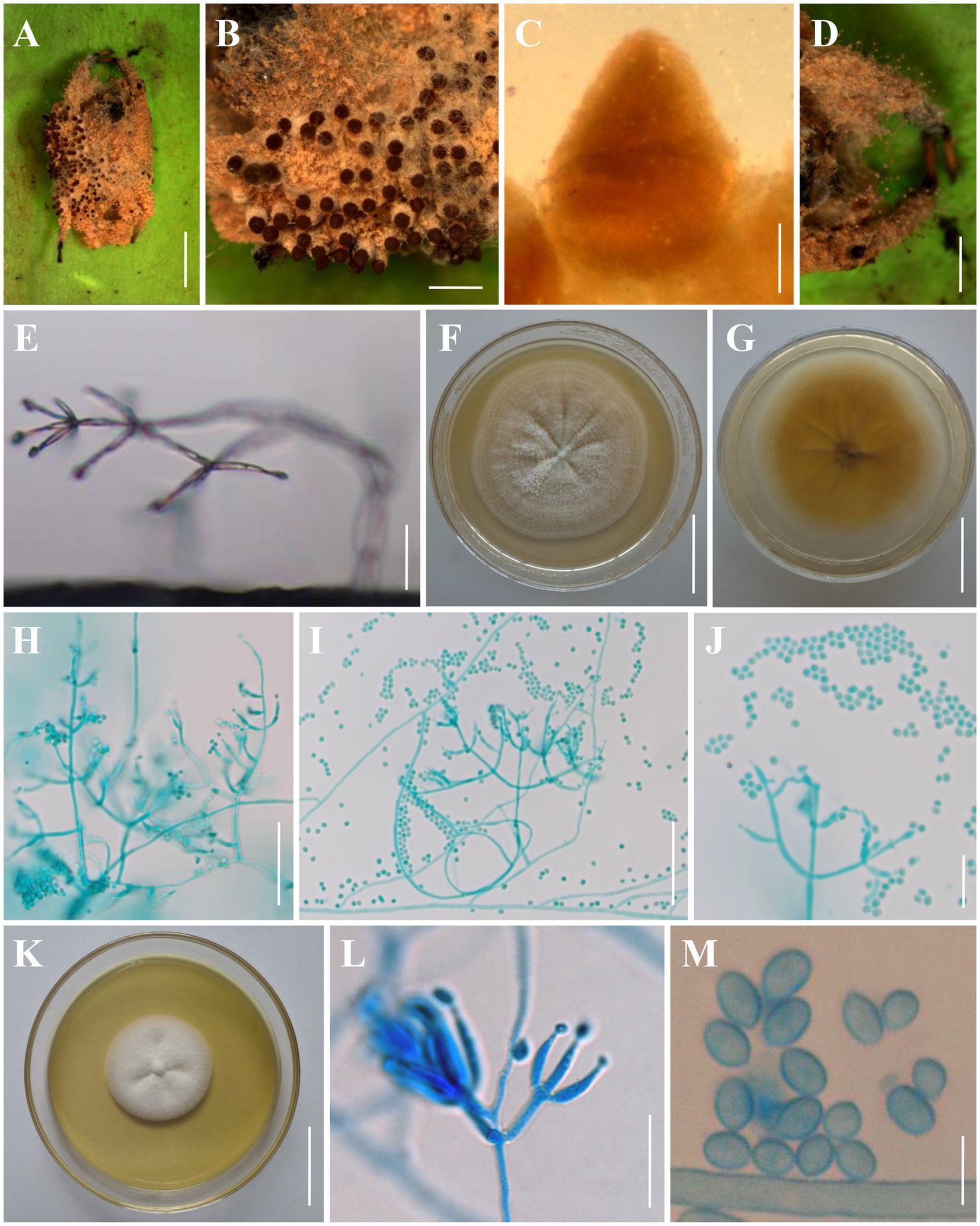
Figure 3. Morphology of Clonostachys chuyangsinensis. (A) Infected spider. (B) Ascomata on the host. (C) Front view of perithecium. (D, E) Conidiogenous structures on the host. (F, G) Colony obverse and reverse on PDA. (H–J) Conidiophores, conidiogenous cells, and conidia on PDA. (K) Colony obverse on CMA. (L) Conidiophores, conidiogenous cells, and conidia on CMA. (M) Conidia on CMA. Scale bars: (A) = 3 mm; (B,D) = 1 mm; (C) = 100 μm; (E,J,L) = 20 μm; (F,G,K) = 30 mm; (H,I) = 50 μm; (M) = 5 μm.
MycoBank number 843885.
Etymology: named after Chu Yang Sin National Park, where this species was first discovered.
Type: Vietnam, Dak Lak Province, Chu Yang Sin National Park (12°29’N, 108°43′E, 1659 m above sea level), on a spider on the underside of a leaf, October 22, 2017, collected by Yuan-Bing Wang (holotype: YHH 896; ex-type: YFCC 896).
Description: Sexual morph: Ascomata on a brown spider, perithecial, solitary or densely crowded in groups, subglobose to oval, (280–)290–380(−400) × (240–)260–330(−340) μm (n = 30), collapsing laterally when dry, pale brown when fresh, becoming dark brown to nearly black when dry, not changing color in 3% KOH or in lactic acid; surface smooth. Asci and ascospores not observed. Asexual morph: Infected spider host covered with a dense brown mycelial mat. Hyphae branched, septate, hyaline, smooth. Conidiophores verticillium-like; phialides divergent in whorls of 2–5 or single from lower levels, generally slightly tapering toward the tip, (5–)5.6–28.3(−36) × (1–)1.4–3.6(−4) μm (n = 30). Conidia smooth-walled, hyaline, subglobose to ellipsoid, (2–)2.5–4.6(−4.8) × (2–)2.4–3.5(−4) μm (n = 30). Colonies on PDA reached 28–32 mm in diameter after 7 days at 25°C, white, circular; reverse pale to light orange (5–6A3–4). Colony surface white powdery to granulose because of the conidiophores and conidial masses; aerial mycelium sparsely produced or absent. Conidiophores monomorphic, verticillate, arising from the agar surface or from the sparse aerial mycelium; stipes (20–)40–130(−150) μm long, (2–)2.5–4(−5) μm wide at the base (n = 50); primary branches divergent, forming independent side-branches; terminal branches and phialides divergent or adpressed; terminal phialides flask-shaped, or cylindrical but narrowing in the upper part, (4.5–)5.5–44.2(−60) × (1.2–)1.5–3.8(−4) μm (n = 50). Conidia in white imbricate columns, smooth-walled, hyaline, subglobose to ellipsoid, (2.2–)2.4–4.7(−5) × (1.5–)1.8–3.5(−3.8) μm (n = 50). Setae not observed. Colonies on CMA reached 25–30 mm in diameter after 7 days at 25°C, white, circular; reverse pale yellowish (1-2A3). Colony surface white powdery due to conidial masses, cottony to felty due to aerial mycelium. Conidiophores monomorphic, verticillate, arising from the agar surface or from the sparse aerial mycelium; stipes (20–)30–145(−160) μm long, (2–)2.5–4(−4.5) μm wide at the base (n = 50); primary branches divergent, forming independent side-branches; terminal branches and phialides divergent or adpressed; terminal phialides flask-shaped, or cylindrical but narrowing in the upper part, (4.5–)5.5–44.2(−50) × (1.2–)1.5–3.8(−4.2) μm (n = 50). Conidia in white imbricate columns, smooth-walled, hyaline, subglobose to ellipsoid, ovoid, (2–)2.2–5(−5.5) × (1.5–)2–3.5(−4) μm (n = 50). Setae not observed.
Distribution: Chu Yang Sin National Park, Dak Lak Province, Vietnam; Kunming City, Yunnan Province, China.
Additional materials examined: China, Yunnan Province, Kunming City, Wild Duck Forest Park (25°13’N, 102°87′E, 2100 m above sea level), from soil on the forest floor, August 20, 2018, Yao Wang (living culture: YFCC 895); China, Yunnan Province, Kunming City, Songming County, Dashao Village (25°24’N, 102°55′E, 2697 m above sea level), from Ophiocordyceps highlandensis, August 25, 2018, De-Xiang Tang (living culture: YFCC 8591) (Zhao et al., 2021).
Notes: Morphologically, C. chuyangsinensis resembles the phylogenetically sister species C. candelabrum. The shape and size of the conidia and the colony color of C. chuyangsinensis among other morphological features have been observed in C. candelabrum. However, C. chuyangsinensis can be distinguished from C. candelabrum by its long phialides ((4.5–)5.5–44.2(−50) × (1.2–)1.5–3.8(−4.2) μm). Both morphological study and phylogenetic analyses of combined ITS, nrLSU, TUB2, and TEF1 sequence data support that this fungus is a distinct species in the genus Clonostachys.
Clonostachys kunmingensis H. Yu & Y. Wang, sp. nov. Figure 4.
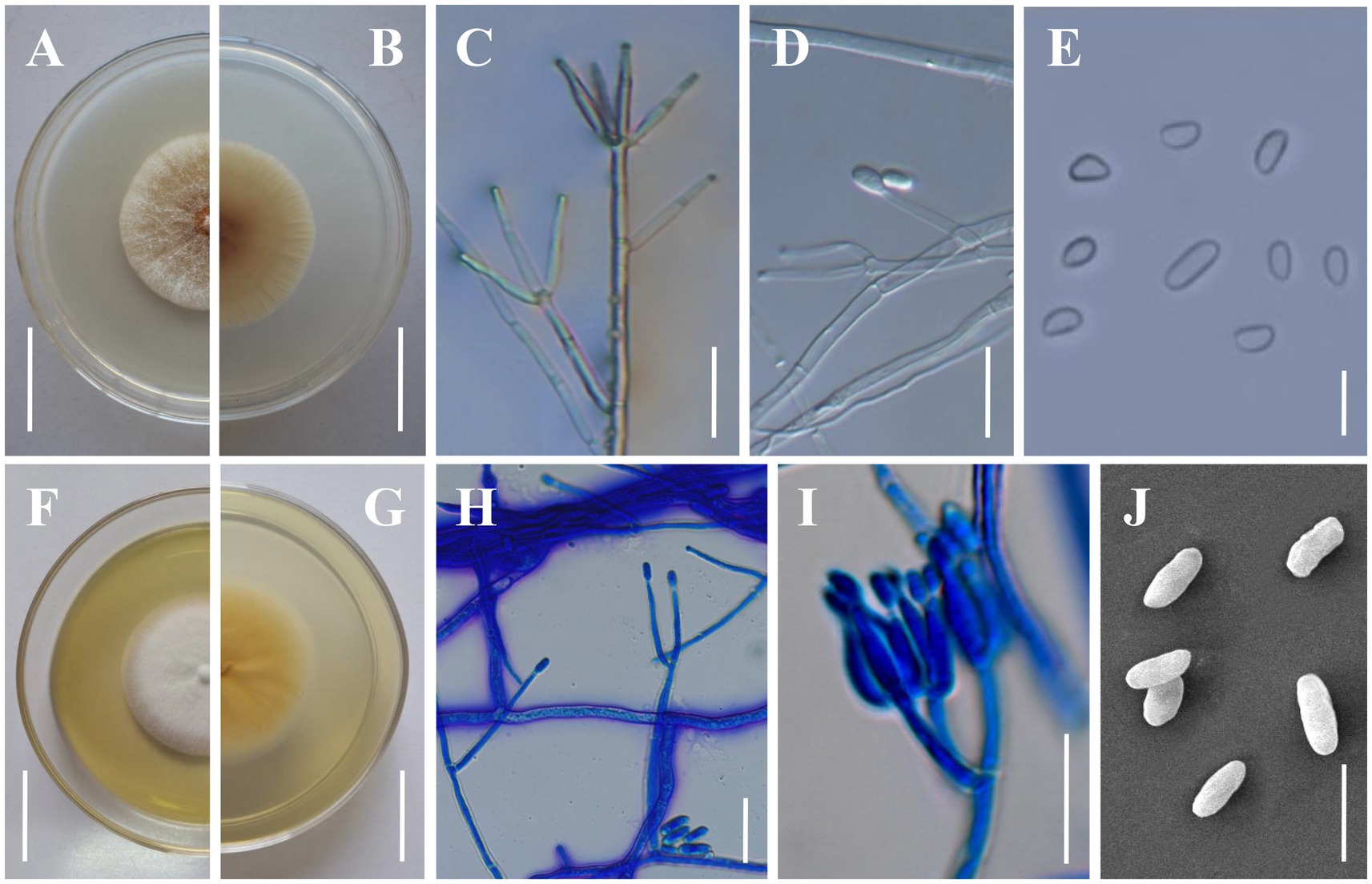
Figure 4. Morphology of Clonostachys kunmingensis. (A,B) Colony obverse and reverse on PDA. (C,D) Verticillium-like primary conidiophores on PDA. (E) Conidia from secondary conidiophores. (F,G) Colony obverse and reverse on CMA. (H) Verticillium-like primary conidiophores on CMA. (I) Secondary conidiophores on CMA. (J) Conidia from primary conidiophores. Scale bars: (A,B,F,G) = 20 mm; (C,D,H,I) = 20 μm; (E,J) = 10 μm.
MycoBank number 843886.
Etymology: named after the location Kunming City where the species was collected.
Type: China, Yunnan Province, Kunming City, Wild Duck Forest Park (25°13’N, 102°87′E, 2100 m above sea level), from soil on the forest floor, August 10, 2019, Yao Wang (holotype: YHH 898, dried specimen; ex-type: YFCC 898).
Description: Sexual morph: Undetermined. Asexual morph: Colonies on PDA reaching 32–35 mm in diameter after 7 days at 25°C, pale yellow (4A2–3), circular; reverse pale orange (5A2–3). Colony surface cottony to felty due to aerial mycelium. Conidiophores dimorphic. Primary conidiophores verticillium-like, arising from the agar surface or from the sparse aerial mycelium; (80–)120–260(−380) μm high, stipes (20–)60–140(−230) μm long, (2–)3.5–5(−5.5) μm wide at the base (n = 50), sometimes with short side branches arising from the upper part; phialides divergent, in whorls of 2–6, sometimes singly from lower levels, (14.2–)19.1–36.4(−52.6) × (2–)2.5–3.5(−3.9) μm (n = 50), straight, cylindrical, slightly tapering toward the tip. Secondary conidiophores penicillate, solitary to gregarious, with divergent branching penicilli; bi-to quarter-verticillate, (15–)30–100(−125) μm long, (3–)3.5–5(−5.5) μm wide at the base (n = 50); penicillus 90–145 μm high, typically with two primary branches, divergent, terminating in moderately divergent metulae and adpressed phialides; phialides divergent or adpressed, in whorls of 2–6, almost cylindrical tapering in the upper part, straight to slightly curved, (5.6–)8.0–17.5(−25) μm long, (2–)2.5–3.2(−4) μm wide at the base, (1–)1.2–1.4(−1.6) μm wide near the aperture (n = 50); intercalary phialides rarely observed. Conidial masses on verticillium-like conidiophores small and round collapsing to form whitish, watery masses; conidial masses on penicillate conidiophores inconspicuous, short, and rather thick, columnar, white. Conidia from secondary conidiophores slightly curved, with one slightly flattened side, distally broadly rounded, with laterally displaced hila, (4–)4.2–8.5(−9) × (2.2–)2.5–4(−4.5) μm (n = 100), held in imbricate; conidia from primary conidiophores larger, oblong to cylindrical, frequently less curved, sometimes without a visible hilum, (6–)6.7–11.2(−14) × (2–)2.3–4.5(−4.8) μm (n = 100). Colonies on CMA reaching 25–32 mm in diameter after 7 days at 25°C, white, circular; reverse yellowish white to light yellow (4A2–5). Colony surface white powdery due to conidial masses. Aerial mycelium on CMA not thick, on PDA strongly developed in thick, often erect hyphal strands. Size and shape of Conidiophores, phialides and conidia similar on PDA and CMA.
Additional materials examined: China, Yunnan Province, Kunming City, Songming County, Dashao Village (25°24’N, 102°55′E, 2750 m above sea level), from soil on the forest floor, August 24, 2019, Yao Wang (living culture: YFCC 892, 967).
Notes: Regarding phylogenetic relationships, C. kunmingensis is closely related to C. rhizophaga and C. chloroleuca and further grouped with C. oblongispora (Figure 2). However, C. kunmingensis can be distinguished from C. rhizophaga and C. chloroleuca by its oblong to cylindrical conidia ((6–)6.7–11.2(−14) × (2–)2.3–4.5(−4.8) μm). Clonostachys kunmingensis consistently showed unpigmented conidial masses, while conidial masses of C. rhizophaga and C. chloroleuca can be greenish or weakly greenish (Moreira et al., 2016). Clonostachys oblongispora differs from C. kunmingensis by its longer conidia ((9–)12.6–13.6–14(−19.8) × (2.6–)3.2–3.6–3.8(−4.2) μm) (Schroers, 2001). Morphologically, C. kunmingensis is similar to C. rosea in terms of the shape and size of the conidiogenous cells and the shape of the conidia (Schroers, 2001). However, our morphological observation revealed some differences between them. Colonies of C. kunmingensis on PDA are pale yellow whereas those of C. rosea are white. Furthermore, conidia from secondary conidiophores of C. kunmingensis ((4–)4.2–8.5(−9) × (2.2–)2.5–4(−4.5) μm) are larger than those of C. rosea ((4.2–)4.8–5.2–5.6(−6.6) × (2–)2.4–2.8–3(−3.4) μm).
Discussion
Clonostachys species are widely distributed and occupy diverse habitats, with various host/substrate associations (see Table 1). The species distribution is cosmopolitan, with the height of known species diversity occurring in tropical regions; the habitat diversity is complicated, with most of the known species having unspecific saprotrophic ability (Schroers, 2001). These known species are commonly found in soils, litter, and dead plant substrata as saprotrophs. They have also been reported as endophytes and epiphytes of living plants (Torcato et al., 2020). Another aspect of the biology of Clonostachys species is their unspecific parasitic ability. Some Clonostachys spp. are known as destructive mycoparasites, with C. rosea and C. rosea f. catenulata being used as biocontrol agents against various ascomycetes, soil-borne hyphomycetes, and basidiomycetes (Schroers, 2001; Chatterton et al., 2008). They are also parasitic to myxomycetes, nematodes, ticks, mollusks, and leafhoppers (Schroers, 2001; Toledo et al., 2006). In this study, we described a novel species, C. chuyangsinensis, which was isolated from a large spider. In fact, Clonostachys species parasitic on spiders have rarely been reported, apart from C. aranearum (Chen et al., 2016). The present study provides new evidence for Clonostachys sp. as an araneopathogenic fungus, thus extending our knowledge of the occurrence and distribution of spider-pathogenic fungi.
Compared with the anamorph of Clonostachys with simple morphological architectures, the teleomorph provided more valuable morphological information to recognize individual Clonostachys species. Schroers (2001) classified the teleomorph in the six distinguished subgenera Astromata, Bionectria, Epiphloea, Myronectria, Uniparietina, and Zebrinella based on stroma morphology, stroma-perithecium wall interface structure, perithecial wall anatomy, habit of the perithecia on the natural substratum, and ascospore ornamentation and septation. Our phylogenetic analyses based on the combined ITS+nrLSU + TUB2 + TEF1 sequences provide additional evidence supporting these morphologically delimited subgenera (Figure 2). It seems that the divisions of six subgenera do not contradict the unity of the entire genus Clonostachys. All taxa of six subgenera are united by the phenotypic characteristics of the anamorph such as penicillate conidiophores, conidia held in imbricate columns, and predominantly more or less curved conidia with mostly laterally displaced hila (Schroers, 2001). Some intraspecific variations in conidiomata, intercalary phialides, conidiophore dimorphism, and conidial mass color have hampered species identification in Clonostachys, but to a certain extent these may reflect subgeneric affinities (Schroers, 2001). In the current study, it should be noted that the phylogenetic trees inferred from the analyses of combined data excluded C. setosa and C. vesiculosa from the six subgenera (Figure 2). The two species should belong to the subgenus Epiphloea based on diagnostic features (Schroers, 2001; Luo and Zhuang, 2010). However, they are distant relatives of Epiphloea spp. from our results (Figure 2). The phenotypic similarities among non-sister species may result from convergent morphological evolution, perhaps due to occupation of similar ecological niches (Bischoff et al., 2009). Therefore, we propose to protect Clonostachys as the genus name for the entire clade, while acknowledging that future studies including more data and taxonomic sampling may introduce new genera to accommodate these subgenera.
The multilocus phylogenetic approach taken in this study of the genus Clonostachys has shed considerable light on this important group of fungi. The results of the present work indicate that the nrLSU sequences provided little valuable information to separate Clonostachys spp., although they were conducive to determining the phylogenomic relationships between Clonostachys and its related genera. In contrast, sequence data for the ITS and protein-coding gene region TUB2 provided good resolution of Clonostachys spp., confirming the results of previous studies (Schroers, 2001; Hirooka and Kobayashi, 2007; Luo and Zhuang, 2010; Chen et al., 2016; Prasher and Chauhan, 2017). Our study also introduced sequence data for the TEF1 gene region. This region requires only two primers and is easily amplified. Although the sequence length of the TEF1 fragment was the shortest among the four loci analyzed in this study, the introns within TEF1 provided the greatest concentration of informative nucleotide variation and degree of phylogenetic resolution for terminal clades in Clonostachys. Additionally, the genetic distances of Clonostachys species for TEF1 were significantly higher than those for ITS and TUB2 (Supplementary Tables S1–S3). Future studies will determine the use of this single locus for the recognition and identification of phylogenetic species in Clonostachys and other fungal species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
YW: conceptualization. YW: methodology, writing—original draft preparation, and formal analysis. YW and RL: software. D-XT and RL: validation. YW, D-XT, Y-BW, CT, and HY: investigation. YW, D-XT, and V-MD: resources. HY: writing—review and editing and funding acquisition. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos 32200013 and 32160005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1117753/full#supplementary-material
Footnotes
References
Abreu, L. M., Moreira, G. M., Ferreira, D., Rodrigues-Filho, E., and Pfenning, L. H. (2014). Diversity of Clonostachys species assessed by molecular phylogenetics and MALDI-TOF mass spectrometry. Fungal Biol. 118, 1004–1012. doi: 10.1016/j.funbio.2014.10.001
Alves, A., Crous, P. W., Correia, A., and Phillips, A. J. L. (2008). Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers 28, 1–13.
Bischoff, J. F., Rehner, S. A., and Humber, R. A. (2009). A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 101, 512–530. doi: 10.3852/07-202
Chatterton, S., Jayaraman, J., and Punja, Z. K. (2008). Colonization of cucumber plants by the biocontrol fungus clonostachys rosea f. catenulata. Biol. Control 46, 267–278. doi: 10.1016/j.biocontrol.2008.02.007
Chen, W. H., Han, Y. F., Liang, J. D., Zou, X., Liang, Z. Q., and Jin, D. C. (2016). A new araneogenous fungus of the genus Clonostachys. Mycosystema 35, 1061–1069. doi: 10.13346/j.mycosystema.150244
Crous, P. W., Shivas, R. G., Quaedvlieg, W., van der Bank, M., Zhang, Y., Summerell, B. A., et al. (2014). Fungal planet description sheets. Persoonia 32, 184–306. doi: 10.3767/003158514X682395
Dao, H. T., Beattie, G. A. C., Rossman, A. Y., Burgess, L. W., and Holford, P. (2016). Four putative entomopathogenic fungi of armoured scale insects on citrus in Australia. Mycol. Prog. 15:47. doi: 10.1007/s11557-016-1188-6
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995
Hirooka, Y., and Kobayashi, T. (2007). Taxonomic studies of nectrioid fungi in Japan. II: the genus Bionectria. Mycoscience 48, 81–89. doi: 10.1007/s10267-006-0331-7
Hyde, K. D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D. J., Jones, E. B. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100, 5–277. doi: 10.1007/s13225-020-00439-5
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lechat, C., and Fournier, J. (2018). Clonostachys spinulosispora (Hypocreales, Bionectriaceae), a new species on palm from French Guiana. Ascomycete.org 10, 127–130. doi: 10.25664/ART-0238
Lechat, C., and Fournier, J. (2020). Two new species of Clonostachys (Bionectriaceae, Hypocreales) from Saül (French Guiana). Ascomycete.org 12, 61–66. doi: 10.25664/ART-0299
Lechat, C., Fournier, J., Chaduli, D., Lesage-Meessen, L., and Favel, A. (2019). Clonostachys sauelensis (Bionectriaceae, Hypocreales), a new species from French Guiana. Ascomycete.org 11, 65–68. doi: 10.25664/ART-0260
Lechat, C., Fournier, J., and Gasch, A. (2020). Clonostachys moreaui (Hypocreales, Bionectriaceae), a new species from the island of Madeira (Portugal). Ascomycete.org 12, 35–38. doi: 10.25664/ART-0295
Lombard, L., van der Merwe, N. A., Groenewald, J. Z., and Crous, P. W. (2015). Generic concepts in Nectriaceae. Stud. Mycol. 80, 189–245. doi: 10.1016/j.simyco.2014.12.002
Luo, J., and Zhuang, W. Y. (2010). Bionectria vesiculosa sp. nov. from Yunnan, China. Mycotaxon 113, 243–249. doi: 10.5248/113.243
Moreira, G. M., Abreu, L. M., Carvalho, V. G., Schroers, H. J., and Pfenning, L. H. (2016). Multilocus phylogeny of Clonostachys subgenus Bionectria from Brazil and description of Clonostachys chloroleuca sp. nov. Mycol. Prog. 15, 1031–1039. doi: 10.1007/s11557-016-1224-6
O’Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/s0953-7562(09)80409-7
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossman, A. Y., Mckemy, J. M., Pardo-Schultheiss, R. A., and Schroers, H.-J. (2001). Molecular studies of the Bionectriaceae using large subunit rDNA sequences. Mycologia 93, 100–110. doi: 10.1080/00275514.2001.12061283
Rossman, A. Y., Seifert, K. A., Samuels, G. J., Minnis, A. M., Schroers, H.-J., Lombard, L., et al. (2013). Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus 4, 41–51. doi: 10.5598/imafungus.2013.04.01.05
Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., et al. (2020). NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020:baaa062. doi: 10.1093/database/baaa062
Schroers, H.-J. (2001). A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud. Mycol. 46, 1–214.
Schroers, H.-J., Samuels, G. J., Seifert, K. A., and Gams, W. (1999). Classification of the mycoparasiteGliocladium roseuminClonostachysas C. Rosea, its relationship toBionectria ochroleuca, and notes on other Gliocladium-like fungi. Mycologia 91, 365–385. doi: 10.1080/00275514.1999.12061028
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Swofford, D. L. (2002). PAUP*. Phylogenetic Analysis using Parsimony (*and other methods), version 4.0b10. Sunderland: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tibpromma, S., Hyde, K. D., McKenzie, E. H. C., Bhat, D. J., Phillips, A. J. L., Wanasinghe, D. N., et al. (2018). Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers. 93, 1–160. doi: 10.1007/s13225-018-0408-6
Toledo, A. V., Virla, E., Humber, R. A., Paradell, S. L., and Lastra, C. C. L. (2006). First record of Clonostachys rosea (Ascomycota: Hypocreales) as an entomopathogenic fungus of Oncometopia tucumana and Sonesimia grossa (Hemiptera: Cicadellidae) in Argentina. J. Invertebr. Pathol. 92, 7–10. doi: 10.1016/j.jip.2005.10.005
Torcato, C., Gonalves, M. F. M., Rodríguez-Gálvez, E., and Alves, A. (2020). Clonostachys viticola sp. nov., a novel species isolated from Vitis vinifera. Int. J. Syst. Evol. Microbiol. 70, 4321–4328. doi: 10.1099/ijsem.0.004286
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cyptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Wang, Y., Wang, Y. R., Han, Y. F., and Liang, Z. Q. (2015). A new thermotolerant species of Taifanglania. Mycosystema 34, 345–349. doi: 10.13346/j.mycosystema.140136
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic)
Zeng, Z. Q., and Zhuang, W. Y. (2017). Three new Chinese records of Hypocreales. Mycosystema 36, 654–662. doi: 10.13346/j.mycosystema.160101
Zhao, P., Luo, J., and Zhuang, W. Y. (2011). Practice towards DNA barcoding of the nectriaceous fungi. Fungal Divers. 46, 183–191. doi: 10.1007/s13225-010-0064-y
Zhao, Z., Zhu, K., Tang, D., Wang, Y., Wang, Y., Zhang, G., et al. (2021). Comparative analysis of mitochondrial genome features among four Clonostachys species and insight into their systematic positions in the order Hypocreales. Int. J. Mol. Sci. 22:5530. doi: 10.3390/ijms22115530
Keywords: Bionectriaceae, molecular systematics, multi-gene phylogeny, morphology, new species
Citation: Wang Y, Tang D-X, Luo R, Wang Y-B, Thanarut C, Dao V-M and Yu H (2023) Phylogeny and systematics of the genus Clonostachys. Front. Microbiol. 14:1117753. doi: 10.3389/fmicb.2023.1117753
Edited by:
Dimitris G. Hatzinikolaou, National and Kapodistrian University of Athens, GreeceReviewed by:
Nalin Nilusha Wijayawardene, Qujing Normal University, ChinaStefania Mirela Mang, University of Basilicata, Italy
Copyright © 2023 Wang, Tang, Luo, Wang, Thanarut, Dao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Yu, hongyu@ynu.edu.cn
 Yao Wang
Yao Wang De-Xiang Tang1,2
De-Xiang Tang1,2 Hong Yu
Hong Yu