-

Figure 1.
Phylogram generated from maximum likelihood analysis based on combined dataset of LSU, SSU, ITS, and TEF1 sequence data. Bootstrap support values for maximum likelihood (ML) equal to or greater than 60% and Bayesian posterior probabilities (BYPP) equal to or greater than 0.97 are given above the nodes. Newly generated sequences are in dark red bold and ex-type isolates are in bold. Lophium mytilinum (AFTOL-ID 1609) and Mytilinidion rhenanum (CBS 135.45) are used as outgroup taxa.
-
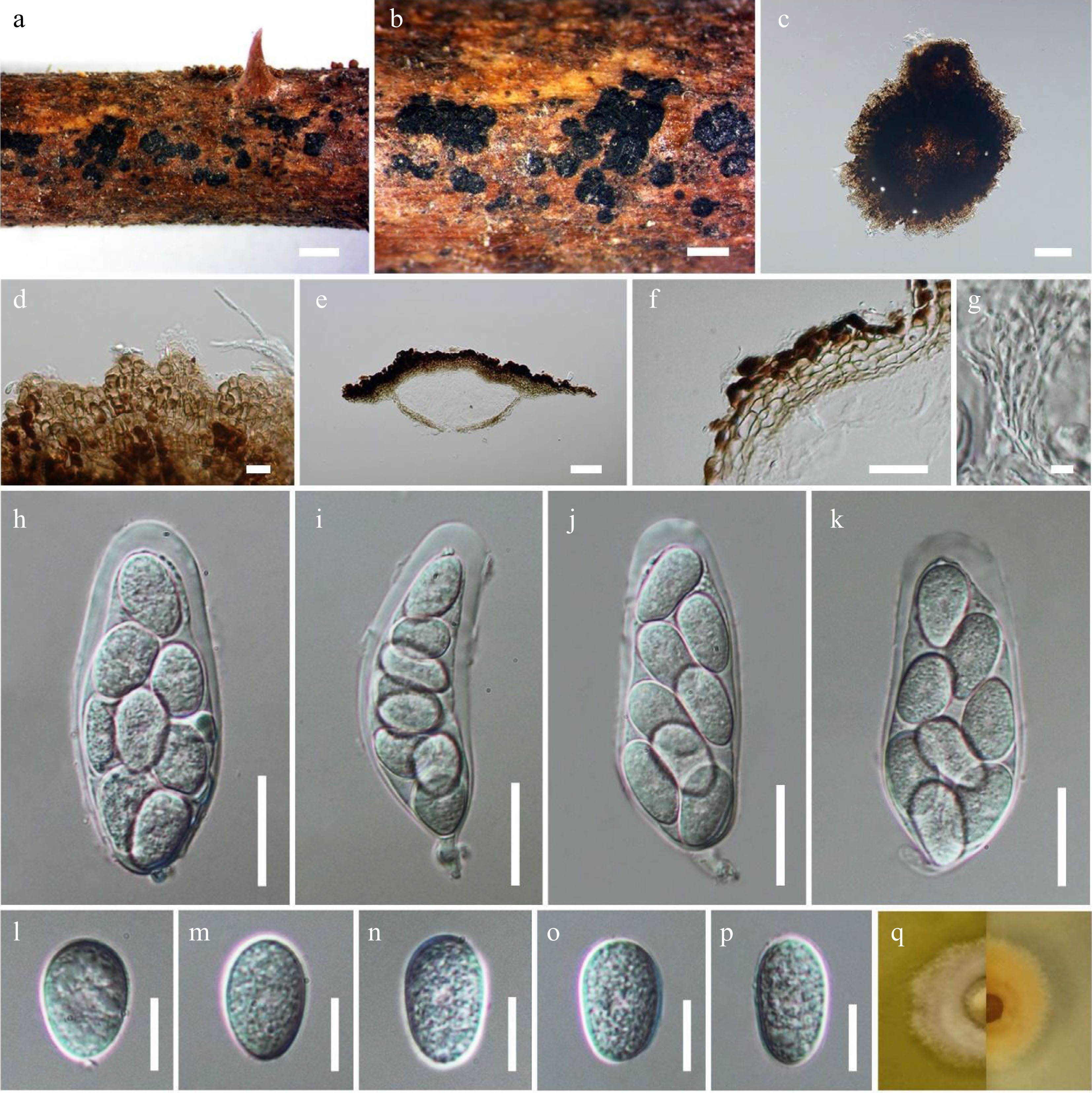
Figure 2.
Muyocopron dipterocarpi (neotype). (a), (b) Superficial ascomata on substrate. (c), (d) Squash mounts showing ascomata walls. (e) Section of ascoma. (f) Peridium. (g) Pseudoparaphyses. (h)−(k). Asci. (l)−(p) Unicellular ascospores. (q) Culture characteristic on MEA. Scale bars: a = 1,000 µm, b = 500 µm, c = 100 µm, e = 50 µm, f, h−k = 20 µm, d, l−p = 10 µm, g = 5 µm.
-
Taxa Strain no.1 GenBank accession numbers2 LSU SSU ITS TEF1 Acrospermum adeanum M133 EU940104 EU940031 EU940180 − Acrospermum compressum M151 EU940084 EU940012 EU940161 − Acrospermum gramineum M152 EU940085 EU940013 EU940162 − Arxiella dolichandrae CBS 138853T KP004477 − KP004449 − Arxiella terrestris CBS 268.65T MH870201 − MH858565 − Dyfrolomyces phetchaburiensis MFLUCC 15-0951T MF615402 MF615403 − − Dyfrolomyces rhizophorae BCC15481 − KF160009 − − Dyfrolomyces rhizophorae JK 5456A GU479799 − − GU479860 Dyfrolomyces thailandica MFLU 16-1173T KX611366 KX611367 − − Dyfrolomyces thamplaensis MFLUCC 15-0635T KX925435 KX925436 − KY814763 Dyfrolomyces tiomanensis NTOU3636T KC692156 KC692155 − KC692157 Leptodiscella africana CBS 400.65T MH870275 − MH858635 − Leptodiscella brevicatenata FMR 10885T FR821311 − FR821312 − Leptodiscella chlamydospora MUCL 28859 FN869567 − FR745398 − Leptodiscella rintelii CBS 144927T LR025181 − LR025180 − Lophium mytilinum AFTOL-ID 1609 DQ678081 DQ678030 − DQ677926 Melomastia maolanensis GZCC 16-0102T KY111905 KY111906 − KY814762 Muyocopron alcornii BRIP 43897T MK487708 − MK487735 MK495956 Muyocopron atromaculans MUCL 34983T MK487709 − MK487736 MK495957 Muyocopron castanopsis MFLUCC 10-0042 − JQ036225 − − Muyocopron castanopsis MFLUCC 14-1108T KU726965 KU726968 MT137784 MT136753 Muyocopron chromolaenae MFLUCC 17-1513T MT137876 MT137881 MT137777 MT136756 Muyocopron chromolaenicola MFLUCC 17-1470T MT137877 MT137882 MT137778 MT136757 Muyocopron coloratum CBS 720.95T MK487710 − NR_160197 MK495958 Muyocopron dipterocarpi MFLU 18-2582 − MW079363 MW063196 − Muyocopron dipterocarpi MFLUCC 14-1103T KU726966 KU726969 MT137785 MT136754 Muyocopron dipterocarpi MFLUCC 17-0075 MH986833 MH986829 MH986837 − Muyocopron dipterocarpi MFLUCC 17-0354 MH986834 MH986830 MH986838 − Muyocopron dipterocarpi MFLUCC 17-0356 MH986835 MH986831 MH986839 − Muyocopron dipterocarpi MFLUCC 17-1464NT OQ861270 OQ861267 OQ832759 OQ856779 Muyocopron dipterocarpi MFLUCC 18-0470 MK348001 MK347890 MK347783 − Muyocopron garethjonesii MFLU 16-2664T KY070274 KY070275 − − Muyocopron geniculatum CBS 721.95T MK487711 − MK487737 MK495959 Muyocopron heveae MFLUCC 17-0066T MH986832 MH986828 MH986836 − Muyocopron laterale CBS 127677 MK487718 − MK487744 MK495965 Muyocopron laterale CBS 141029T MK487712 − MK487738 MK495960 Muyocopron laterale CBS 141033 MK487715 − MK487741 MK495963 Muyocopron laterale CBS 145309 MK487722 − MK487748 MK495969 Muyocopron laterale CBS 145310 MK487719 − MK487745 MK495966 Muyocopron laterale CBS 145311 MK487724 − MK487750 − Muyocopron laterale CBS 145312 MK487725 − MK487751 MK495971 Muyocopron laterale CBS 145313 MK487721 − MK487747 MK495968 Muyocopron laterale CBS 145314 MK487723 − MK487749 MK495970 Muyocopron laterale CBS 145315 MK487720 − MK487746 MK495967 Muyocopron laterale CBS 145316 MK487726 − MK487752 MK495972 Muyocopron laterale CBS 719.95 MK487714 − MK487740 MK495962 Muyocopron laterale FMR 13797 MK874616 − MK874615 MK875803 Muyocopron laterale IMI 324533 MK487713 − MK487739 MK495961 Muyocopron laterale URM 7801 MK487717 − MK487743 − Muyocopron laterale URM 7802 MK487716 − MK487742 MK495964 Muyocopron lithocarpi − MK447738 MK447740 − − Muyocopron lithocarpi MFLU 18-2087 MK347930 MK347821 MK347716 − Muyocopron lithocarpi MFLU 18-2088 MK347931 MK347822 MK347717 − Muyocopron lithocarpi MFLUCC 10-0041 JQ036230 JQ036226 − − Muyocopron lithocarpi MFLUCC 14-1106T KU726967 KU726970 MT137786 MT136755 Muyocopron lithocarpi MFLUCC 16-0962 MK348034 MK347923 − − Muyocopron lithocarpi MFLUCC 17-1465 MT137878 MT137883 MT137779 MT136758 Muyocopron lithocarpi MFLUCC 17-1466 MT137879 MT137884 MT137780 MT136759 Muyocopron lithocarpi MFLUCC 17-1500 MT137880 MT137885 MT137781 MT136760 Muyocopron zamiae CBS 203.71T MK487727 − − MK495973 Mycoleptodiscus endophytica MFLUCC 17-0545T MG646946 MG646978 MG646961 MG646985 Mycoleptodiscus suttonii CBS 141030 MK487729 − − MK495975 Mycoleptodiscus suttonii CBS 276.72T MK487728 − MK487753 MK495974 Mycoleptodiscus terrestris CBS 231.53T MK487730 − MK487754 MK495976 Mycoleptodiscus terrestris IMI 159038 MK487731 − MK487755 MK495977 Mytilinidion rhenanum CBS 135.34 FJ161175 FJ161136 − FJ161092 Neocochlearomyces chromolaenae BCC 68250T MK047514 MK047552 MK047464 MK047573 Neocochlearomyces chromolaenae BCC 68251 MK047515 MK047553 MK047465 MK047574 Neocochlearomyces chromolaenae BCC 68252 MK047516 MK047554 MK047466 MK047575 Neomycoleptodiscus venezuelense CBS 100519T MK487732 − MK487756 MK495978 Palawania thailandensis MFLU 16-1871 KY086494 − MT137788 − Palawania thailandensis MFLUCC 14-1121T KY086493 KY086495 MT137787 − Paramycoleptodiscus albizziae CBS 141320 KX228330 − KX228279 MK495979 Paramycoleptodiscus albizziae CPC 27552T MH878220 − − − Pseudopalawania siamensis MFLUCC 17-1476aT − MT137789 MT137782 MT136752 Pseudopalawania siamensis MFLUCC 17-1476b − MT137790 MT137783 − Setoapiospora thailandica MFLUCC 17-1426T MN638847 MN638851 MN638862 MN648731 1 AFTOL-ID: Assembling the Fungal Tree of Life; BCC: BIOTEC Culture Collection; BRIP: Biosecurity Queensland Plant Pathology Herbarium, Brisbane, Australia; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CPC: Culture collection of Pedro Crous, the Netherlands; FMR: Facultad de Medicina, Reus, Tarragona, Spain; GZCC: Guizhou Culture Collection; IMI: The International Mycological Institute Culture Collections; JK: J. Kohlmeyer; MFLU: the Herbarium of Mae Fah Luang University; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUCL: Belgian Coordinated Collections of Microorganisms; URM: Universidade Federal de Pernambuco; T: ex-type isolates; NT: Neotype.
2 LSU: 28S large subunit of the nrRNA gene; SSU: 18S small subunit of the nrRNA gene; ITS: internal transcribed spacer regions 1 and 2 including 5.8S nrRNA gene; TEF1: partial translation elongation factor 1-α gene.Table 1.
Taxa used in this study and their GenBank accession numbers. New sequences are in bold.
Figures
(2)
Tables
(1)