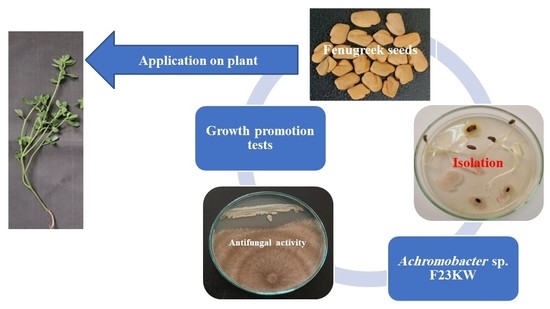
Seed Endophytic Achromobacter sp. F23KW as a Promising Growth Promoter and Biocontrol of Rhizoctonia Root Rot of Fenugreek
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Pathogenic Fungus
2.2. Preparation of R. solani Inoculum
2.3. Isolation of Endophytic Bacterial Microbiome
2.4. In Vitro Evaluation of Endophytic Bacteria against R. solani
2.5. Biochemical Profile of Endophytic Bacteria
2.5.1. Bacterial Inoculum
2.5.2. Culturing Technique
2.5.3. Assay of Hydrolytic Enzymes
2.6. Gas Chromatography-Mass Spectrometry (GC-MS)
2.7. Morphological, Biochemical, and Molecular Identification
2.8. Greenhouse Experiment
2.8.1. Disease Assessment
2.8.2. Growth Parameters
2.8.3. Estimation of Total Phenol
2.8.4. Defense-Related Enzymes
2.9. Field Trial
2.10. Experimental Design and Statistical Analysis
3. Results
3.1. Isolation and Antagonistic Activity of Endophytic Bacteria
3.2. Profile of Biochemical Features
3.3. GC-MS Profile
3.4. Bacterial Identification
3.5. Greenhouse Evaluation
3.5.1. Achromobacter sp. F23KW vis R. solani
3.5.2. Growth Features of Fenugreek Seedlings
3.5.3. Physiological Performance of Fenugreek Seedlings
3.6. Performance of Fenugreek under Field Conditions
3.6.1. Growth and Yield
3.6.2. Disease Progress under Field Conditions
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sarwar, S.; Hanif, M.A.; Ayub, M.A.; Boakye, Y.D.; Agyare, C. Fenugreek. In Medicinal Plants of South Asia; Hanif, M.A., Nawaz, H., Khan, M.M., Byrne, H.J., Eds.; Elsevier: London, UK, 2019; pp. 258–269. [Google Scholar] [CrossRef]
- Chaudhary, S.; Chaudhary, P.S.; Chikara, S.K.; Sharma, M.C.; Iriti, M. Review on fenugreek (Trigonella foenum-graecum L.) and its important secondary metabolite Diosgenin. Not. Bot. Horti Agrobot. 2018, 46, 22–31. [Google Scholar] [CrossRef]
- Parthasarathy, V.A.; Kandinnan, K.; Srinivasan, V. Fenugreek. In Organic Spices; New India Publishing Agencies: New Delhi, India, 2008; p. 694. [Google Scholar]
- Singh, R.B.; Smail, M.M.A.; Rai, R.H.; Maheshwari, A.; Verma, N.; Isaza, A. Effects of fenugreek seeds on cardiovascular diseases and other chronic diseases. In Functional Foods and Nutraceuticals in Metabolic and NonCommunicable Diseases; Singh, R.B., Watanabe, S., Isaza, A.A., Eds.; Academic Press: London, UK, 2022; pp. 399–410. [Google Scholar] [CrossRef]
- Acharya, K.; Chakraborty, N.; Chatterjee, S.; Basu, S.K. Fungal Diseases of Fenugreek. Fenugreek Special Issue. Am. J. Soc. Issues Huminities 2014, 176, 171–185. [Google Scholar]
- Meena, R.D.; Meena, R.S.; Sharma, Y.K.; Meena, S.S.; Meena, N.L. Response of fenugreek varieties against Rhizoctonia solani, causing root rot. Int. J. Seed Spices 2020, 10, 73–75. [Google Scholar]
- Al-Surhanee, A.A.; Afzal, M.; Bouqellah, N.A.; Ouf, S.A.; Muhammad, S.; Jan, M.; Kaleem, S.; Hashem, M.; Alamri, S.; Abdel Latef, A.A.H.; et al. The Antifungal Activity of Ag/CHI NPs against Rhizoctonia solani linked with tomato plant health. Plants 2021, 10, 2283. [Google Scholar] [CrossRef]
- Yadav, S.L.; Ghasolia, R.P.; Yadav, R. Reaction of fenugreek varieties against Rhizoctonia solani, causing root rot disease. Int. J. Curr. Microbiol. App. Sci. 2019, 8, 2878–2882. [Google Scholar] [CrossRef]
- Rani, N.; Hegde, Y.R. Survey for the Incidence of Root Rot/Wilt of Fenugreek in Northern Karnataka, India. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 1564–1569. [Google Scholar] [CrossRef]
- Al-Askar, A.A.; Ghoneem, K.M.; Hafez, E.E.; Saber, W.I.S. A case study in saudi arabia: Biodiversity of maize seed-borne pathogenic fungi in relation to biochemical, physiological, and molecular characteristics. Plants 2022, 11, 829. [Google Scholar] [CrossRef]
- Mohamadpoor, M.; Amini, J.; Ashengroph, M.; Azizi, A. Evaluation of biocontrol potential of Achromobacter xylosoxidans strain CTA8689 against common bean root rot. Physiol. Mol. Plant Pathol. 2022, 117, 101769. [Google Scholar] [CrossRef]
- Saber, W.I.A.; El-Naggar, N.E.; AbdAl-Aziz, S.A. Bioconversion of lignocellulosic wastes into organic acids by cellulolytic rock phosphate-solubilizing fungal isolates grown under solid-state fermentation conditions. Res. J. Microbiol. 2010, 5, 1–20. [Google Scholar] [CrossRef]
- Al-Askar, A.A.; Rashad, E.M.; Ghoneem, K.M.; Mostafa, A.A.; Al-Otibi, F.O.; Saber, W.I.A. Discovering Penicillium polonicum with high-lytic capacity on Helianthus tuberosus tubers: Oil-based preservation for mold management. Plants 2021, 10, 413. [Google Scholar] [CrossRef]
- Bailey, M.J.; Beily, P.; Poutanen, K. Interlaboratory testing and methods for assay of xylanase activity. J. Biotechnol. 1992, 23, 257–270. [Google Scholar] [CrossRef]
- Abdelwahed, N.A.M.; El-Naggar, N.E.A.; Saber, W.I.A. Factors and correlations controlling cellulase-free xylanase production by Streptomyces halstedii NRRL B-1238 in submerged culture. Aust. J. Basic Appl. Sci. 2011, 5, 45–53. [Google Scholar]
- Gurumallesh, P.; Alagu, K.; Ramakrishnan, B.; Muthusamy, S. A systematic reconsideration on proteases. Int. J. Biol. Macromol. 2019, 128, 254–267. [Google Scholar] [CrossRef]
- Shen, C.R.; Chen, Y.S.; Yang, C.J.; Chen, J.K.; Liu, C.L. Colloid chitin azure is a dispersible, low-cost substrate for chitinase measurements in a sensitive, fast, reproducible assay. J. Biomol. Screen. 2010, 15, 213–217. [Google Scholar] [CrossRef]
- Singh, R.S.; Singh, T.; Pandey, A. Microbial Enzymes—An Overview. In Biomass, Biofuels, Biochemicals: Advances in Enzyme Technology, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–40. [Google Scholar] [CrossRef]
- Saber, W.I.; Ghoneem, K.M.; Rashad, Y.M.; Al-Askar, A.A. Trichoderma Harzianum WKY1: An indole acetic acid producer for growth improvement and anthracnose disease control in sorghum. Biocontrol Sci. Technol. 2017, 27, 654–676. [Google Scholar] [CrossRef]
- ISTA. International Rules for Seed Testing, Rules 1999; Seed Science and Technology: Zürich, Switzerland, 1999; Volume 24, pp. 1–335. [Google Scholar]
- Bai, Z.H.; Zhang, H.X.; Qi, H.Y.; Peng, X.W.; Li, B.J. Pectinase production by Aspergillus niger using wastewater in solid state fermentation for eliciting plant disease resistance. Bioresour. Technol. 2004, 95, 49–52. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Garrity, G.M.; Brenner, D.J.; Krieg, N.R.; Staley, J.T. (Eds.) Bergey’s Manual of Systematic Bacteriology, Volume Two: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria; Springer: New York, NY, USA, 2005; ISBN 9780387241456. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Malik, C.P.; Singh, M.B. Plant Enzymology and Histo Enzymology; Kalyani Publishers: New Delhi, India, 1980; p. 286. [Google Scholar]
- Blainski, A.; Lopes, G.C.; de Mello, J.C.P. Application and analysis of the folin ciocalteu method for the determination of the total phenolic content from Limonium brasiliense L. Molecules 2013, 18, 6852–6865. [Google Scholar] [CrossRef]
- Beaudoin-Egan, L.D.; Thorpe, T.A. Tyrosine and phenylalanine ammonia lyase activities during shoot inhibition in tobacco callus cultures. Plant Physiol. 1985, 78, 438–441. [Google Scholar] [CrossRef]
- Seleim, M.A.; Abo-Elyousr, K.A.; Mohamed, A.A.A.; Al-Marzoky, H.A. Peroxidase and polyphenoloxidase activities as biochemical markers for biocontrol efficacy in the control of tomato bacterial wilt. Plant Physiol. Pathol. 2014, 2, 2. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Behrendt, U.; Ahmad, P.; Berg, G. Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 2017, 8, 199. [Google Scholar] [CrossRef]
- Padder, S.A.; Rather, R.A.; Bhat, S.A.; Shah, M.D.; Baba, T.R.; Mubarak, N.M. Dynamics, phylogeny and phyto-stimulating potential of chitinase synthesizing bacterial root endosymbiosiome of North Western Himalayan Brassica rapa L. Sci. Rep. 2022, 12, 6742. [Google Scholar] [CrossRef]
- Glassner, H.; Zchori-Fein, E.; Yaron, S.; Sessitsch, A.; Sauer, U.; Compant, S. Bacterial niches inside seeds of Cucumis melo L. Plant Soil 2018, 422, 101–113. [Google Scholar] [CrossRef]
- El-Hersh, M.S.; Saber, W.I.; El-Fadaly, H.A. Amino acids associated with optimized alkaline protease production by Bacillus subtilis ATCC 11774 using statistical approach. Biotechnology 2014, 13, 252–262. [Google Scholar] [CrossRef]
- Moussa, Z.; Darwish, D.B.; Alrdahe, S.S.; Saber, W.I. Innovative artificial-intelligence-based approach for the biodegradation of feather keratin by Bacillus paramycoides, and cytotoxicity of the resulting amino acids. Front. Microbiol. 2021, 12, 731262. [Google Scholar] [CrossRef]
- Berndt, D.; Spahrbier, D. Batteries. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH: Weinheim, Germany, 2005. [Google Scholar] [CrossRef]
- Ismael, A.; Gevorgyan, A.; Skrydstrup, T.; Bayer, A. Renewable solvents for palladium-catalyzed carbonylation reactions. Org. Process Res. Dev. 2020, 24, 2665–2675. [Google Scholar] [CrossRef]
- Özçakmak, S.; Dervisoglu, M.; Yilmaz, A. Antifungal activity of lemon balm and sage essential oils on the growth of ochratoxigenic Penicillium verrucosum. Afr. J. Microbiol. Res. 2012, 6, 3079–3084. [Google Scholar] [CrossRef]
- Uwineza, P.A.; Urbaniak, M.; Bryła, M.; Stȩpień, Ł.; Modrzewska, M.; Waśkiewicz, A. In vitro effects of lemon balm extracts in reducing the growth and mycotoxins biosynthesis of Fusarium culmorum and F. proliferatum. Toxins 2022, 14, 355. [Google Scholar] [CrossRef] [PubMed]
- Said, S.M.; Hammam, M.A.; Abd-El Kader, S.K. Insecticidal activity against the greater wax moth (Galleria mellonella L.) and chemical composition of five plant essential oils. Menoufia J. Plant Prot. 2019, 4, 145–161. [Google Scholar] [CrossRef]
- Martin, J.H.; Knevel, A.M. Gas chromatographic method of moisture determination. J. Pharm. Sci. 1965, 54, 1464–1467. [Google Scholar] [CrossRef]
- Dowanol, P.M. Propylene Glycol Methyl Ether; 1-Methoxy-2-Propanol. Available online: https://www.ataman-chemicals.com/en/products/dipropylene-glycol-monomethyl-ether-2176.html (accessed on 19 August 2022).
- Bosen, S.F.; Bowles, W.A.; Ford, E.A.; Perlson, B.D. Antifreezes. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH: Weinheim, Germany, 2000. [Google Scholar] [CrossRef]
- Kinnunen, T.; Koskela, M. Antibacterial and antifungal properties of Propylene glycol, Hexylene glycol, and 1,3-Butylene glycol in vitro. Acta Derm.-Venereol. 1991, 71, 148–150. [Google Scholar]
- Nalawade, T.M.; Bhat, K.; Sogi, S.H. Bactericidal activity of propylene glycol, glycerine, polyethylene glycol 400, and polyethylene glycol 1000 against selected microorganisms. J. Int. Soc. Prevent. Communit. Dent. 2015, 5, 114–119. [Google Scholar] [CrossRef]
- Sakkas, H.; Papadopoulou, C. Antimicrobial Activity of basil, oregano, and thyme essential oils. J. Microbiol. Biotechnol. 2017, 27, 429–438. [Google Scholar] [CrossRef]
- Khalil, N.; Ashour, M.; Fikry, S.; Naser, A.; Salama, O. Future Journal of Pharmaceutical Sciences Chemical composition and antimicrobial activity of the essential oils of selected Apiaceous fruits. Future J. Pharm. Sci. 2018, 4, 88–92. [Google Scholar] [CrossRef]
- Núñez-Carmona, E.; Abbatangelo, M.; Sberveglieri, V. Characterization and Analysis of Volatile Fingerprint of 13 Different Commercial Essential Oils with GC-MS and Chemical Gas Sensors. Preprints 2018, 2018090568. [Google Scholar] [CrossRef]
- Yigit, F.; Dikilitas, M. Control of Fusarium wilt of tomato by combination of fluorescent Pseudomonas, non-pathogen Fusarium and Trichoderma harzianum T22 in greenhouse conditions. J. Plant Pathol. 2007, 6, 159–163. [Google Scholar] [CrossRef]
- De Boer, M.; Van der Sluis, L.; Van Loon, L.C.; Bakker, A.H.M.P. Combining fluorescent Pseudomonas spp. strain to enhance suppression of Fusarium wilt of radish. Eur. J. Plant Pathol. 1999, 105, 201–210. [Google Scholar] [CrossRef]
- Joe, M.M.; Islam, M.R.; Karthikeyan, B.; Bradeepa, K.; Sivakumaar, P.K.; Sa, B. Resistance responses of rice to rice blast fungus after seed treatment with the endophytic Achromobacter xylosoxidans AUM54 strain. Crop Prot. 2012, 42, 141–148. [Google Scholar] [CrossRef]
- Wu, W.; Chen, W.; Liu, S.; Wu, J.; Zhu, Y.; Qin, L.; Zhu, B. Beneficial relationships between endophytic bacteria and medicinal plants. Front. Plant Sci. 2021, 12, 646146. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Kumar, A. Characterization of novel plant growth promoting endophytic bacterium Achromobacter xylosoxidans from wheat plant. Microb. Ecol. 2009, 58, 179–188. [Google Scholar] [CrossRef]
- Karthikeyan, B.; Joe, M.M.; Islam, R.; Sa, T. ACC deaminase containing diazotrophic endophytic bacteria ameliorate salt stress in Catharanthus roseus through reduced ethylene levels and induction of antioxidative defense systems. Symbiosis 2012, 56, 77–86. [Google Scholar] [CrossRef]
- Zhang, Y.F.; He, L.Y.; Chen, Z.J.; Wang, Q.Y.; Qian, M.; Sheng, X.F. Characterization of ACC deaminase producing endophytic bacteria isolated from copper tolerant plants and their potential in promoting the growth and copper accumulation of Brassica napus. Chemosphere 2011, 83, 57–62. [Google Scholar] [CrossRef]
- Jhuma, T.A.; Rafeya, J.; Sultana, S.; Rahman, M.T.; Karim, M.M. Isolation of endophytic salt-tolerant plant growth-promoting Rhizobacteria from Oryza sativa and evaluation of their plant growth-promoting traits under salinity stress condition. Front. Sustain. Food Syst. 2021, 5, 687531. [Google Scholar] [CrossRef]
- Hungria, M.; Campo, R.J.; Souza, E.M.; Pedrosa, F.O. Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 2010, 331, 413–425. [Google Scholar] [CrossRef]
- Chandra, D.; Sharma, A.K. Field evaluation of consortium of bacterial inoculants producing ACC deaminase on growth, nutrients and yield components of rice and wheat. J. Crop Sci. Biotechnol. 2021, 24, 293–305. [Google Scholar] [CrossRef]
- Schmidt, C.S.; Mrnka, L.; Lovecká, P.; Frantík, T.; Fenclová, M.; Demnerová, K.; Vosátka, M. Bacterial and fungal endophyte communities in healthy and diseased oilseed rape and their potential for biocontrol of Sclerotinia and Phoma disease. Sci. Rep. 2021, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Sultana, A.; Shupta, S.A.; Chakraborty, S.; Khokon, M.A.R. Evaluation of foliar spraying of Bacillus subtilis and Achromobacter xylosoxidans for management of bacterial leaf blight (BLB) of rice under field condition. Bangladesh J. Plant Pathol. 2020, 36, 39–48. [Google Scholar]
- Romero, F.M.; Rossi, F.R.; Gárriz, A.; Carrasco, P.; Ruíz, O.A. A bacterial endophyte from apoplast fuids protects canola plants from diferent pathogens via antibiosis and induction of host resistance. Phytopathology 2019, 109, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yuan, J.; Raza, W.; Shen, Q.; Huang, Q. Biocontrol traits and antagonistic potential of Bacillus amyloliquefaciens strain NJZJSB3 against Sclerotinia sclerotiorum, a causal agent of canola stem rot. J. Microbiol. Biotechnol. 2014, 24, 1327–1336. [Google Scholar] [CrossRef]
- Kamal, M.M.; Lindbeck, K.D.; Savocchia, S.; Ash, G.J. Biological control of Sclerotinia stem rot of canola using antagonistic bacteria. Plant Pathol. 2015, 64, 1375–1384. [Google Scholar] [CrossRef]






| Isolate | Xylanase (U) | FPase (U) | PGase (U) | Proteinase (U) | Chitinase (U) | IAA (µg/g) | Final pH |
|---|---|---|---|---|---|---|---|
| F1KW | 61.92 ± 5.01 | 3.20 ± 0.97 | 2.91 ± 0.27 | ND | 0.19 ± 0.04 | 242.8 ± 4.8 | 8.4 ± 0.5 |
| F3KW | 60.61 ± 4.12 | 3.14 ± 0.55 | 1.49 ± 0.64 | ND | 0.57 ± 0.12 | 255.1 ± 9.5 | 8.4 ± 0.3 |
| F9KW | 55.78 ± 3.41 | 2.89 ± 0.61 | 2.53 ± 0.81 | ND | 1.10 ± 0.12 | 119.3 ± 8.2 | 8.4 ± 0.4 |
| F7KW | 67.63 ± 3.57 | 3.50 ± 0.45 | 2.34 ± 0.93 | ND | 2.30 ± 0.91 | 205.8 ± 5.9 | 8.3 ± 0.2 |
| F14KW | 65.44 ± 2.56 | 3.39 ± 1.01 | 2.30 + 0.34 | ND | 0.53 ± 0.27 | 242.8 ± 4.9 | 8.2 ± 0.6 |
| F17KW | ND | ND | 2.03 ± 0.54 | ND | ND | 144.0 ± 7.3 | 8.2 ± 0.2 |
| F22KW | 59.73 ± 3.39 | 3.09 ± 0.99 | 1.76 ± 0.09 | 8.47 ± 1.33 | ND | 170.0 ± 6.1 | 8.3 ± 0.3 |
| F23KW | 70.71 ± 4.47 | 3.50 ± 0.83 | 3.10 ± 1.00 | 7.50 ± 1.96 | 6.20 ± 1.29 | 353.9 ± 5.6 | 8.5 ± 0.4 |
| F19KW | 65.44 ± 1.95 | 3.39 ± 0.92 | 1.53 ± 0.82 | ND | 1.10 ± 0.99 | 218.1 ± 5.8 | 8.5 ± 0.1 |
| F25KW | 67.63 ± 4.85 | 3.66 ± 0.31 | 1.72 ± 0.39 | 7.23 ± 2.47 | 4.80 ± 1.38 | 157.6 ± 3.9 | 8.4 ± 0.5 |
| Peak No. | Retention Time (min) | Compound | Formula | Molecular Weight | Retention Index | Area | Height | Area Sum, % |
|---|---|---|---|---|---|---|---|---|
| 1 | 6.516 | Dipropylene glycol monomethyl ether | C7H16O3 | 148.200 | 1034 | 562,758 | 269,890 | 6.43 |
| 2 | 7.386 | 2-Propanol, 1,1’-oxybis- | C6H14O3 | 134.174 | 1018 | 2,395,965 | 793,699 | 27.37 |
| 3 | 7.580 | Propane, 1,2-dimethoxy- | C5H12O2 | 104.148 | 859 | 1,984,213 | 979,929 | 22.66 |
| 4 | 7.627 | Methane, diethoxy- | C5H12O2 | 104.148 | 843 | 3,256,992 | 1,456,742 | 37.20 |
| 5 | 7.924 | 2-Butanol, 3,3′-oxybis- | C8H18O3 | 162.227 | 1089 | 555,580 | 339,089 | 6.35 |
| Taxonomic Feature | Strain F23KW |
|---|---|
| General features | |
| Gram reaction | − |
| H2S production | − |
| Lipid hydrolysis | + |
| Gelatin hydrolysis | − |
| Citrate utilization | − |
| Phenylalanine deamination | − |
| Indole test | + |
| Indole-3-acetic acid | + |
| Carbohydrate fermentation | |
| Xylose | + |
| Maltose | + |
| Lactose | − |
| Maltose | + |
| Mannitol | − |
| Sucrose | + |
| Mannose | − |
| Fructose | + |
| Tolerance to NaCl (% w/v) | Up to 3.0 |
| pH range of growth | 6–9.0 |
| Enzymatic activity | |
| Chitinase | + |
| Xylanase | + |
| Cellulase | + |
| Pectinase | + |
| Proteinase | + |
| Urease | − |
| Oxidase | + |
| Catalase | + |
| Nitrate reductase | + |
| Treatment | Pre-Emergence Damping Off, % * | Post-Emergence Damping Off, % * | Plant Survival, % * | |
|---|---|---|---|---|
| Infected | P | 25.0 a | 17.2 a | 57.8 c |
| PBC | 14.2 b | 10.2 b | 75.6 b | |
| PBS | 13.2 b | 10.8 b | 76.0 b | |
| PBCS | 11.8 b | 8.20 bc | 80.0 b | |
| PF | 13.2 b | 10.2 b | 76.6 b | |
| Noninfected | Control | 3.40 c | 3.20 c | 93.4 a |
| BC | 5.60 c | 5.00 bc | 89.4 a | |
| BS | 5.80 c | 4.00 c | 90.2 a | |
| BCS | 5.40 c | 3.80 c | 90.8 a | |
| Treatment | Plant Length (cm) | Leaves Number per Plant | Plant Weight (g) | |||
|---|---|---|---|---|---|---|
| Shoot | Root | Fresh | Dry | |||
| Infected | P | 7.3 c | 3.8 c | 4.2 c | 334.0 c | 47.0 d |
| PBC | 13.8 b | 5.5 ab | 7.4 a | 504.0 ab | 90.0 ab | |
| PBS | 17.5 a | 5.3 ab | 6.0 ab | 548.0 ab | 92.0 ab | |
| PBCS | 17.7 a | 6.4 a | 7.0 ab | 542.0 ab | 104.0 ab | |
| PF | 12.1 b | 4.9 bc | 5.6 bc | 412.0 bc | 61.8 cd | |
| Noninfected | Control | 14.0 b | 4.8 bc | 6.2 ab | 495.8 ab | 83.8 bc |
| BC | 14.8 ab | 6.0 ab | 7.2 ab | 516.0 ab | 90.6 ab | |
| BS | 17.3 a | 6.2 ab | 7.0 ab | 552.0 ab | 93.0 ab | |
| BCS | 17.8 a | 6.4 a | 7.6 a | 576.0 a | 112.6 a | |
| Treatment | Total Phenols (100 mg/g) | Polyphenol Oxidase (U) | Peroxidase (U) | |
|---|---|---|---|---|
| Infected | P | 392.68 d | 13.0 c | 17.33 d |
| PBC | 405.02 cd | 28.0 b | 1588.67 a | |
| PBS | 435.09 bcd | 28.2 b | 566.67 cd | |
| PBCS | 482.17 b | 40.3 a | 936.67 bc | |
| PF | 407.03 cd | 29.0 b | 513.34 cd | |
| Noninfected | Control | 459.13 bc | 23.0 bc | 788.67 bc |
| BC | 440.09 bcd | 29.0 b | 1045.33 abc | |
| BS | 484.17 b | 42.0 a | 1361.33 ab | |
| BCS | 565.32 a | 49.0 a | 766.00 bc | |
| Treatment | Shoot Length (cm) | Root Length (cm) | Branches Number/Plant | Leaves Number/Plant | Plant Fresh Weight (g) | Plant Dry Weight (g) | Pods Number/Plant | Weight of 100 Dry Pods | No. Seeds/Plant | Total Yield (ton/h) |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | 39.8 b | 14.4 d | 3.6 c | 37.0 b | 17.39 c | 3.01 b | 11.1 b | 26.20 d | 294.33 e | 0.294 e |
| BC | 49.6 a | 18.1 bc | 5.8 ab | 57.4 a | 29.37 ab | 4.11 ab | 18.8 ab | 33.52 b | 408.67 c | 0.414 c |
| BS | 51.6 a | 19.8 ab | 5.6 ab | 49.4 ab | 31.96 ab | 4.46 a | 20.0 ab | 33.48 b | 440.00 b | 0.438 b |
| BCS | 55.6 a | 20.6 a | 6.4 a | 66.2 a | 36.16 a | 4.77 a | 25.0 a | 38.54 a | 505.33 a | 0.473 a |
| Fungicide | 48.0 ab | 16.6 c | 4.8 bc | 50.6 ab | 27.01 b | 3.97 ab | 18.0 ab | 29.90 c | 371.67 d | 0.359 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashad, E.M.; Shaheen, D.M.; Al-Askar, A.A.; Ghoneem, K.M.; Arishi, A.A.; Hassan, E.S.A.; Saber, W.I.A. Seed Endophytic Achromobacter sp. F23KW as a Promising Growth Promoter and Biocontrol of Rhizoctonia Root Rot of Fenugreek. Molecules 2022, 27, 5546. https://doi.org/10.3390/molecules27175546
Rashad EM, Shaheen DM, Al-Askar AA, Ghoneem KM, Arishi AA, Hassan ESA, Saber WIA. Seed Endophytic Achromobacter sp. F23KW as a Promising Growth Promoter and Biocontrol of Rhizoctonia Root Rot of Fenugreek. Molecules. 2022; 27(17):5546. https://doi.org/10.3390/molecules27175546
Chicago/Turabian StyleRashad, Ehsan M., Dalia M. Shaheen, Abdulaziz A. Al-Askar, Khalid M. Ghoneem, Amr Abker Arishi, El Sayed A. Hassan, and WesamEldin I. A. Saber. 2022. "Seed Endophytic Achromobacter sp. F23KW as a Promising Growth Promoter and Biocontrol of Rhizoctonia Root Rot of Fenugreek" Molecules 27, no. 17: 5546. https://doi.org/10.3390/molecules27175546