Metabolomics-Guided Analysis of the Biocatalytic Conversion of Sclareol to Ambradiol by Hyphozyma roseoniger
Abstract
:1. Introduction
2. Results
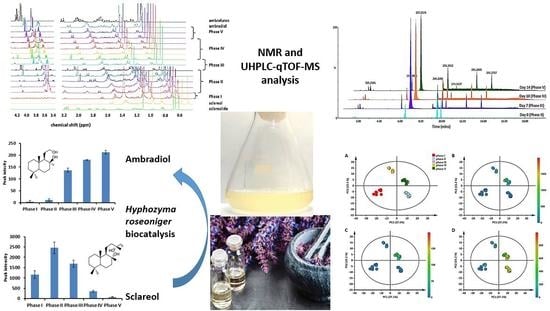
2.1. NMR Characterization
2.2. UHPLC-MS Analysis and Semi-Targeted Metabolomics Analysis of the Biocatalytic Conversion of Sclareol to Ambradiol by H. roseoniger
2.3. OPLS-DA Modeling and Metabolite Annotation of LC–MS Analysed Compounds
3. Discussion
4. Materials and Methods
4.1. Growth of Hyphozyma roseoniger
4.2. Batch Culture in Liquid Medium
4.3. Harvesting of Cells
4.4. Metabolite Extraction, Concentration, and Sample Preparation
4.5. Sample Analysis and Data Acquisition
4.5.1. Ultra-High Performance Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry (UHPLC–qTOF–MS) Analyses
4.5.2. Nuclear Magnetic Resonance (NMR) Analyses
4.6. Data Handling: Pre-Processing and Pre-Treatment
4.7. Multivariate Data Analyses and Statistical Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sultana, N. Microbial biotransformation of bioactive and clinically useful steroids and some salient features of steroids and biotransformation. Steroids 2018, 136, 76–92. [Google Scholar] [CrossRef]
- Wohlgemuth, R. Asymmetric biocatalysis with microbial enzymes and cells. Curr. Opin. Microbiol. 2010, 13, 283–292. [Google Scholar] [CrossRef]
- Allendes, J.A.; Bustos, D.A.; Del, A.; Pacciaroni, V.; Sosa, V.E.; Bustos, D.A. Microbial functionalization of (–)-ambroxide by filamentous fungi. Biocatal. Biotransform. 2011, 29, 83–86. [Google Scholar] [CrossRef]
- Choi, J.; Han, S.; Kim, H. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef]
- Hegazy, M.F.; Mohamed, T.A.; Elshamy, A.I.; Mohamed, A.H.; Mahalel, U.A.; Reda, E.H.; Shaheen, A.M.; Tawfik, W.A. Microbial biotransformation as a tool for drug development based on natural products from mevalonic acid pathway: A review. J. Adv. Res. 2015, 6, 17–33. [Google Scholar] [CrossRef] [Green Version]
- Cheetham, P.S.J. The use of biotransformations for the production of flavours and fragrances. Trends Biotechnol. 1993, 11, 478–488. [Google Scholar] [CrossRef]
- Vandamme, E.J. Bioflavours and fragrances via fungi and their enzymes. Fungal Divers. 2003, 13, 153–166. [Google Scholar]
- Ncube, E.N.; Steenkamp, L.H.; Dubery, I.A. Ambrafuran (AmbroxTM) synthesis from natural plant product precursors. Molecules 2020, 25, 3851. [Google Scholar] [CrossRef] [PubMed]
- Schempp, F.M.; Drummond, L.; Buchhaupt, M.; Schrader, J. Microbial cell factories for the production of terpenoid flavor and fragrance compounds. J. Agric. Food Chem. 2018, 66, 2247–2258. [Google Scholar] [CrossRef]
- Ząbek, A.; Klimek-Ochab, M.; Jawień, E.; Młynarz, P. Biodiversity in targeted metabolomics analysis of filamentous fungal pathogens by 1H NMR-based studies. World J. Microbiol. Biotechnol. 2017, 33, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sevastos, A.; Kalampokis, I.F.; Panagiotopoulou, A.; Pelecanou, M.; Aliferis, K.A. Implication of Fusarium graminearum primary metabolism in its resistance to benzimidazole fungicides as revealed by 1H NMR metabolomics. Pestic. Biochem. Physiol. 2018, 148, 50–61. [Google Scholar] [CrossRef]
- Ezekiel, C.N.; Oyedele, O.A.; Kraak, B.; Ayeni, K.I.; Sulyok, M.; Houbraken, J.; Krska, R. Fungal diversity and mycotoxins in low moisture content ready-to-eat foods in Nigeria. Front. Microbiol. 2020, 11, 615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, N.D.; Gowen, C.M.; Lussier, F.X.; Cautha, S.C.; Mahadevan, R.; Martin, V.J.J. Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics. Microb. Cell Fact. 2015, 14, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spevacek, A.R.; Benson, K.H.; Bamforth, C.W.; Slupsky, C.M. Beer metabolomics: Molecular details of the brewing process and the differential effects of late and dry hopping on yeast purine metabolism. J. Inst. Brew. 2016, 122, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA sequencing and LC–MS-based metabolomics to evaluate the effects of live yeast on rumen function in beef cattle. Animals 2019, 9, 28. [Google Scholar] [CrossRef] [Green Version]
- Farbood, M.I.; Willis, B.J. Process for Producing Diol and Furan and Microorganism Capable of Same. European Patent EP0204009A1, 14 March 1989. [Google Scholar]
- Steenkamp, L.H.; Taka, M. Process for the Production of Ambrafuran. U.S. Patent US20100248316A1, 30 September 2010. [Google Scholar]
- Ratcliffe, R.G.; Shachar-Hill, Y. Revealing metabolic phenotypes in plants: Inputs from NMR analysis. Biol. Rev. 2005, 80, 27–43. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S. Metabolomics: The principles and potential applications to transplantation. Am. J. Transplant. 2005, 5, 2814–2820. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; De Vos, R.C.H.; Vervoort, J. Metabolomics technologies and metabolite identification. Trends Anal. Chem. 2007, 26, 855–866. [Google Scholar] [CrossRef]
- Ncube, E.N.; Mathiba, K.; Steenkamp, L.H.; Dubery, I.A. Gas chromatographic profiling of the biocatalytic conversion of sclareol to ambradiol by Hyphozyma roseoniger. Biocatal. Biotransform. 2022, 40, 1–5. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. A chemometrics toolbox based on projections and latent variables. J. Chemom. 2014, 28, 332–346. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Mongwe, J. Development of an Optimized Process for Commercial Production of (–)-Ambrafuran. Master’s Dissertation, University of Witwatersrand, Johannesburg, South Africa, 2016. [Google Scholar]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Yamamoto, H.; Yamaji, H.; Abe, Y.; Harada, K.; Waluyo, D.; Fukusaki, E.; Kondo, A.; Ohno, H.; Fukuda, H. Dimensionality reduction for metabolome data using PCA, PLS, OPLS, and RFDA with differential penalties to latent variables. Chemom. Intell. Lab. Syst. 2009, 98, 136–142. [Google Scholar] [CrossRef]
- Cheetham, P.S.J. Screening for novel biocatalysts. Enzyme Microbiol. Technol. 1987, 9, 194–213. [Google Scholar] [CrossRef]
- Igarashi, K.; Takizawa, S.; Higaki, N.; Hagihara, H. Microorganism and Method for Producing Dodecahydro-3a, 6, 6, 9a-tetramethyl naphtho[2, 1-b]furan Intermediate, Using the Novel Microorganism. U.S. Patent US8084237B2, 27 December 2011. [Google Scholar]
- Farbood, M.I.; Morris, J.A.; Downey, A.E. Process for Producing Diol and Lactone and Microorganisms Capable of Same. U.S. Patent US4970163A, 13 November 1990. [Google Scholar]
- Farooq, A.; Tahara, S. Biotransformation of two cytotoxic terpenes, α-santonin and sclareol by Botrytis cinerea. Zeitschrift für Naturforschung C 2000, 55, 713–717. [Google Scholar] [CrossRef]
- Martins, M.P.; Ouazzani, J.; Arcile, G.; Jeller, A.H.; de Lima, J.P.F.; Seleghim, M.H.R.; Oliveira, A.L.L.; Debonsi, H.M.; Venâncio, T.; Yokoya, N.S.; et al. Biohydroxylation of (−)-Ambrox®, (−)-Sclareol, and (+)-Sclareolide by whole cells of Brazilian marine-derived fungi. Mar. Biotechnol. 2015, 17, 211–218. [Google Scholar] [CrossRef]
- Díez, D.; Sanchez, J.; Rodilla, J.; Rocha, P.; Mendes, R.; Paulino, C.; Marcos, I.; Basabe, P.; Urones, J. Microbial hydroxylation of sclareol by Rhizopus stolonifer. Molecules 2005, 10, 1005–1009. [Google Scholar] [CrossRef] [Green Version]
- Bustos Crescentino, D.; Pacciaroni, A.; Bustos, D.A.; Sosa, V. Conversion of (-)-ambroxide by whole cells of Fusarium verticillioides (Hypocreales, Nectriaceae). Int. J. Appl. Microbiol. Biotechnol. Res. 2016, 4, 1–6. [Google Scholar] [CrossRef]
- Jassbi, A.R.; Zamanizadehnajari, S.; Azar, P.A.; Tahar, S. Antibacterial diterpenoids from Astragalus brachystachys. Zeitschrift für Naturforschung C 2002, 57, 1016–1021. [Google Scholar] [CrossRef]
- Kouzi, S.A.; McChesney, J.D. Microbial models of mammalian metabolism: Fungal metabolism of the diterpene sclareol by Cunninghamella species. J. Nat. Prod. 1991, 54, 483–490. [Google Scholar] [CrossRef]
- Kouzi, S.A.; McChesney, J.D. Hydroxylation and glucoside conjugation in the microbial metabolism of the diterpene sclareol. Xenobiotica 1991, 21, 1311–1323. [Google Scholar] [CrossRef]
- Hanson, J.R.; Hitchcock, P.B.; Nasir, H.; Truneh, A. The biotransformation of the diterpenoid, sclareol, by Cephalosporium aphidicola. Phytochemistry 1994, 36, 903–906. [Google Scholar] [CrossRef]
- Zerbe, P.; Chiang, A.; Yuen, M.; Hamberger, B.; Hamberger, B.; Draper, J.A.; Britton, R.; Bohlmann, J. Bifunctional cis-abienol synthase from Abies balsamea discovered by transcriptome sequencing and its implications for diterpenoid fragrance production. J. Biol. Chem. 2012, 287, 12121–12131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Brink, J.M.; van Gorcom, R.F. Oxdoreductase from flamentous fungi, DNA coding therefor and cells transformed with said DNA. U.S. Patent US5801024A, 1 September 1998. [Google Scholar]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450 a Success Story. Genome Biol. 2000, 1, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munro, A.W.; Girvan, H.M.; Mason, A.E.; Dunford, A.J.; McLean, K.J. What makes a P450 tick? Trends Biochem. Sci. 2013, 38, 140–150. [Google Scholar] [CrossRef]
- Hieda, T.; Mikami, Y.; Obi, Y.; Kisaki, T. Microbial transformation of cis-abienol. Agric. Biol. Chem. 1982, 46, 2249–2255. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Yao, Q.; Hua, D.; Qin, J. Comparative proteomic analyses of Hyphozyma roseonigra ATCC 20624 in response to sclareol. Braz. J. Microbiol. 2019, 50, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Van der Werf, M.J.; de Bont, J.A.M.; Leak, D.J. Opportunities in microbial biotransformation of monoterpenes. In Biotechnology of Aroma Compounds; Berger, R.G., Babel, W., Blanch, H.W., Cooney, C.L., Enfors, S.O., Eriksson, E.L., Fiechter, A., Klibanov, A.M., Primrose, S.B., Rogers, P.L., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; Volume 55, pp. 147–177. [Google Scholar]






| # | Metabolite | Molecular Formula | Rt (min) | MW (g/mol) | [M+H]+ | Probability | Fold Change |
|---|---|---|---|---|---|---|---|
| Day 0 vs. Day 3 | |||||||
| 1 | Reynosin I | C15H20O | 2.49 | 248.32 | 249.1443 | 9.53 × 104 | 0.59 |
| Reynosin II | C15H20O | 4.50 | 248.32 | 249.1436 | 1.02 × 107 | 0.26 | |
| Day 3 vs. Day 14 | |||||||
| 3 | Sclareolide I | C16H26O2 | 5.63 | 250.38 | 251.1942 | 1.07 × 103 | 421.61 |
| 4 | Ambradiol (product ion) | C16H30O2 (C16H29O) | 6.51 | 254.41 | (* 237.2172) | 8.80 × 104 | 51.18 |
| 5 | Sclareolide II | C16H26O2 | 9.10 | 250.38 | 251.1958 | 5.18 × 105 | 3.00 |
| 6 | Sclareol (degradation product) | C20H36O2 (C20H33) | 9.54 | 308.51 | (** 273.2523) | 2.52 × 105 | 5.76 |
| 7 | epi-Sclareol (degradation product) | C20H36O2 (C20H33) | 10.00 | 308.51 | (** 273.2531) | 6.41 × 106 | 11.21 |
| 8 | cis/trans-Abienol | C20H34O | 11.86 | 290.50 | 291.2597 | 6.21 × 105 | 40.39 |
| 9 | cis/trans-Abienol | C20H34O | 13.28 | 290.50 | 291.2596 | 4.00 × 104 | 52.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ncube, E.N.; Steenkamp, P.A.; van der Westhuyzen, C.W.; Steenkamp, L.H.; Dubery, I.A. Metabolomics-Guided Analysis of the Biocatalytic Conversion of Sclareol to Ambradiol by Hyphozyma roseoniger. Catalysts 2022, 12, 55. https://doi.org/10.3390/catal12010055
Ncube EN, Steenkamp PA, van der Westhuyzen CW, Steenkamp LH, Dubery IA. Metabolomics-Guided Analysis of the Biocatalytic Conversion of Sclareol to Ambradiol by Hyphozyma roseoniger. Catalysts. 2022; 12(1):55. https://doi.org/10.3390/catal12010055
Chicago/Turabian StyleNcube, Efficient N., Paul A. Steenkamp, Chris W. van der Westhuyzen, Lucia H. Steenkamp, and Ian A. Dubery. 2022. "Metabolomics-Guided Analysis of the Biocatalytic Conversion of Sclareol to Ambradiol by Hyphozyma roseoniger" Catalysts 12, no. 1: 55. https://doi.org/10.3390/catal12010055