Abstract
Strawberry is a small fruit crop with high economic value. Anthracnose caused by Colletotrichum spp. poses a serious threat to strawberry production, particularly in warm and humid climates, but knowledge of pathogen populations in tropical and subtropical regions is limited. To investigate the diversity of infectious agents causing strawberry anthracnose in Taiwan, a disease survey was conducted from 2010 to 2018, and Colletotrichum spp. were identified through morphological characterization and multilocus phylogenetic analysis with internal transcribed spacer, glyceraldehyde 3-phosphate dehydrogenase, chitin synthase, actin, beta-tubulin, calmodulin, and the intergenic region between Apn2 and MAT1-2-1 (ApMAT). Among 52 isolates collected from 24 farms/nurseries in Taiwan, a new species, Colletotrichum miaoliense sp. nov. (6% of all isolates), a species not previously known to be associated with strawberry, Colletotrichum karstii (6%), and three known species, Colletotrichum siamense (75%), Colletotrichum fructicola (11%), and Colletotrichum boninense (2%), were identified. The predominant species C. siamense and C. fructicola exhibited higher mycelial growth rates on potato dextrose agar and caused larger lesions on wounded and non-wounded detached strawberry leaves. Colletotrichum boninense, C. karstii, and C. miaoliense only caused lesions on wounded leaves. Understanding the composition and biology of the pathogen population will help in disease management and resistance breeding.
Similar content being viewed by others
Introduction
Strawberry (Fragaria × ananassa Duch.) is a popular small fruit crop with high economic and nutritive value. Strawberry is in high demand globally. From 2008 to 2018, the annual worldwide cultivation of strawberries increased from approximately 400 to 483 thousand hectares1. Although strawberries are native to temperate regions, they can also be grown in tropical and subtropical regions (sometimes under high-altitude conditions). The land areas devoted to strawberry cultivation in Colombia, Peru, Guatemala, Bolivia, and Taiwan in 2018 were 1,482 ha, 1,453 ha, 690 ha, 522 ha, and 506 ha, respectively1.
Anthracnose caused by Colletotrichum spp. is a serious threat to strawberry production, especially in warm and humid climates2. Rain-splashed conidia of Colletotrichum spp. serve as the major inoculum causing epidemics of strawberry anthracnose disease3. After landing on the plant surface, the conidia germinate, form appressoria, then penetrate the epidermal cells4. Colletotrichum spp. can infect various strawberry tissues, causing black spots or irregular spots on leaves, sunken black spots or necrosis lesions on petioles, stolons, and fruits, and wilting of the whole plant due to crown rot2. Under high humidity, concentric rings of acervuli with orange conidial masses can be observed on necrotic tissues. In the US state of Florida, anthracnose causes the death of up to 80% of seedlings in the nursery and yield losses of over 50% in the field2. In Taiwan, strawberry seedlings are propagated from March to September, and the high temperature, high humidity and heavy rainfall during this period provide a suitable environment for epidemics. From 2010 to 2016, anthracnose crown rot caused the loss of 30–40% of seedlings and ~ 20% of plants after transplanting5.
Colletotrichum spp. have traditionally been classified based on the shape of the conidia and appressorium, the presence of a seta or perithecium, and culture characteristics6,7. Using these criteria, early studies reported C. acutatum, C. gloeosporioides, and C. fragariae as strawberry anthracnose pathogens2,8. However, Colletotrichum spp. share similar features, and morphological characteristics can be influenced by environmental factors including culture media, light, and temperature9,10,−11. Therefore, a polyphasic approach based on morphology and genetic characteristics was proposed for identification of Colletotrichum species9. A combination of multiple gene sequences, including internal transcribed spacer (ITS), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), chitin synthase (CHS-1), actin (ACT), beta-tubulin (TUB2), calmodulin (CAL), and the intergenic region between Apn2 and MAT1-2-1 (ApMAT), can provide more molecular features to resolve different species in a Colletotrichum species complex12,13. Through multilocus sequence analysis coupled with morphological characterization, recent studies have identified many additional Colletotrichum species associated with strawberry, namely C. acutatum, C. fioriniae, C. godetiae, C. nymphaeae, C. salicis and C. simmondsii (C. acutatum species complex), C. aenigma, C. changpingense, C. fructicola, C. gloeosporioides, C. siamense and C. theobromicola (syn. C. fragariae) (C. gloeosporioides species complex) and C. boninense (C. boninense species complex)6,12,13,14,15,16,17,18.
Although strawberry is of great economic importance in Taiwan and anthracnose has become more destructive in the past decade, the pathogen population in Taiwan has not been systematically investigated. The causal agents of strawberry anthracnose were previously reported to be C. gloeosporioides19, C. dematium, C. fragariae, and C. acutatum (Plant Protection Information System; https://otserv2.tactri.gov.tw/ppm/), but information about the isolation, pathogenicity, morphology, and sequences of these species is not sufficient for species identification. Recently, based on analysis of multiple gene sequences, we identified C. siamense as the pathogen causing anthracnose crown rot5. To provide accurate information for disease control and resistance breeding, in this study we aimed to reveal the population composition of the infectious agents associated with strawberry anthracnose in Taiwan. Samples collected from the major strawberry-producing areas of Taiwan from 2010 to 2018 were subjected to morphological and multi-gene phylogenetic analyses. To further understand the in vitro and in planta aggressiveness of different Colletotrichum spp. at different temperatures, multiple representative isolates of each species were tested for mycelial growth rates in an artificial medium as well as the ability to cause lesions on wounded or non-wounded strawberry leaves. Since population analysis of Colletotrichum spp. causing strawberry anthracnose has only been reported for species from the UK14 and China [Anhui, Hainan (only one isolate), Hebei, Hubei, Liaoning, Shandong, and Zhejiang Provinces and Beijing and Shanghai cities]16,17,18, which, with the exception of Hainan, are geographical regions located at higher latitudes (30–53°N) relative to Taiwan (24.5°N), this study will provide insights into the biology of strawberry anthracnose disease in subtropical regions.
Results
Molecular identification and phylogenetic analysis
Colletotrichum spp. isolates were first identified at the species complex level. Among 52 Colletotrichum spp. isolates sampled from the major strawberry-producing areas of Taiwan, 45 (86.5%) isolates belonged to the C. gloeosporioides species complex, 4 (7.7%) belonged to the C. boninense species complex, and 3 (5.8%) belonged to the C. acutatum species complex (Table 1).
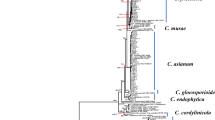
To further analyze the C. acutatum species complex, 3 isolates together with 40 reference isolates, including the outgroup C. orchidophilum (CBS 632.80), were used to construct phylogenetic trees with five gene sequences (ITS, GAPDH, CHS-1, ACT, and TUB2) (Table 1 and Supplementary Table S1) following Damm et al.12 and Fu et al.20. The final data matrix contained a total of 1,821 characters with gaps (ITS: 1–540, GAPDH: 541–799, CHS-1: 800–1,081, ACT: 1,082–1,329, TUB2: 1,330–1,821), of which 237 characters were parsimony informative, 174 parsimony uninformative, and 1,410 constant. After 2,000,000 generations of topological convergence via Bayesian inference (BI) analysis, 2,378 trees were obtained. The first 25% of the trees were discarded, representing the burn-in phase of the analyses, and the remaining trees were used to calculate the Bayesian posterior probabilities in the majority rule consensus tree (Fig. 1). The maximum likelihood (ML) analysis resulted in a best scoring RAxML tree with a final optimized likelihood value of − 6,726.174303. The most parsimonious tree resulted from the maximum parsimony (MP) analysis received tree length = 692, consistency index (CI) = 0.714, and retention index (RI) = 0.843. All three isolates (ML1040, ML1042, ML1794) were grouped in a distinct clade with significant statistical support in the multilocus phylogenetic analysis (1/100/100, BI/ML/MP) (Fig. 1) and the single gene trees of GAPDH, CHS-1, and TUB2 (Supplementary Fig. S1). This clade was distinct from all other known species, and is herein described as a new species, C. miaoliense sp. nov.
A Bayesian inference phylogenetic tree of the C. acutatum species complex. The phylogenetic tree was built using concatenated sequences of the ITS and the GAPDH, ACT, CHS-1 and TUB2 genes. Bayesian inference (BI) posterior values above 0.9 and bootstrap support values from maximum likelihood (ML) and maximum parsimony (MP) above 70% are shown at each node (BI/ML/MP). C. orchidophilum CBS 632.80 was used as the outgroup. *Indicates the ex-type strains. Strains isolated in this study are shown in bold.
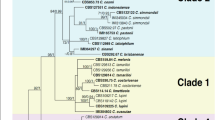
To analyze the phylogeny of the C. boninense species complex, six gene sequences (ITS, GAPDH, CHS-1, ACT, TUB2, and CAL) from 4 isolates together with 31 reference isolates, including the outgroup sequence of C. gloeosporioides (IMI 356878), were used to construct phylogenetic trees (Table 1 and Supplementary Table S1). The final data matrix contained a total of 2,363 characters with gaps (ITS: 1–558, GAPDH: 559–852, CHS-1: 853–1,132, ACT: 1,133–1,411, TUB2: 1,412–1,914, CAL: 1,915–2,363), of which 365 characters were parsimony informative, 407 parsimony uninformative, and 1,591 constant. After 1,187,000 generations of topological convergence via BI analysis, 492 trees were obtained. The first 25% of the trees were discarded, representing the burn-in phase of the analyses, and the remaining trees were used to calculate the Bayesian posterior probabilities in the majority rule consensus tree (Fig. 2). The ML analysis resulted in a best scoring RAxML tree with a final optimized likelihood value of − 10,025.941645. The most parsimonious tree resulted from the MP analysis received tree length = 1,281, CI = 0.774, and RI = 0.839. In single gene trees of GAPDH, CHS-1, and TUB2 and the multilocus phylogenetic tree, three isolates (ML351, 442, 1792) clustered with strong statistical support in the clade containing the type strain CGMCC 3.14194 and other related isolates of C. karstii (Fig. 2; single gene trees not shown). In single gene trees of GAPDH and CAL and the multilocus phylogenetic tree, the isolate ML521 clustered with strong statistical support in the clade containing the type strain CBS 123755 and other related isolates of C. boninense (Fig. 2; single gene trees not shown).
A Bayesian inference phylogenetic tree of the C. boninense species complex. A phylogram was built using concatenated sequences of the ITS and the GAPDH, ACT, CHS-1, TUB2 and CAL genes. Bayesian inference (BI) posterior values above 0.9 and bootstrap support values from maximum likelihood (ML) and maximum parsimony (MP) above 70% are shown at each node (BI/ML/MP). C. gloeosporioides IMI 356878 was used as the outgroup. *Indicates the ex-type strains. Strains isolated in this study are shown in bold.
To identify species in the C. gloeosporioides species complex, a combination of seven gene sequences (ITS, GAPDH, CHS-1, ACT, TUB2, CAL and ApMAT) from 45 isolates together with 47 reference isolates, including the outgroup sequence of C. boninense (CBS 123755), were used to construct phylogenetic trees (Table 1 and Supplementary Table S1). The final data matrix contained a total of 3,571 characters with gaps (ITS: 1–553, GAPDH: 554–808, CHS-1: 809–1,051, ACT: 1,052–1,325, TUB2: 1,326–2,028, CAL: 2,029–2,728, ApMAT: 2,729–3,571), of which 655 characters were parsimony informative, 735 parsimony uninformative, and 2,181 constant. After 6,574,000 generations of topological convergence via BI analysis, 9,864 trees were obtained. The first 25% of the trees were discarded, representing the burn-in phase of the analyses, and the remaining trees were used to calculate the Bayesian posterior probabilities in the majority rule consensus tree (Fig. 3). The ML analysis resulted in a best scoring RAxML tree with a final optimized likelihood value of − 18,514.217014. The most parsimonious tree resulted from the MP analysis received tree length = 2,400, CI = 0.722, and RI = 0.876. In single gene trees of TUB2, CAL, and ApMAT and the multilocus phylogenetic tree, 39 isolates clustered with strong statistical support in the clade containing the type strain CBS 130417 and other related isolates of C. siamense (Fig. 3; single gene trees not shown). The 39 isolates formed a subclade with a high support value (1/100/98, BI/ML/MP) (Fig. 3). In single gene trees of GAPDH, ACT, TUB2, CAL, and ApMAT and the multilocus phylogenetic tree, six isolates clustered with strong statistical support in the clade containing the type strain CBS 130416 and other related isolates of C. fructicola (Fig. 3; single gene trees not shown).
A Bayesian inference phylogenetic tree of the C. gloeosporioides species complex. The phylogenetic tree was built using concatenated sequences of the ITS, ApMAT, and the GAPDH, ACT, CHS-1, TUB2, CAL genes. Bayesian inference (BI) posterior values above 0.9 and bootstrap support values from maximum likelihood (ML) and maximum parsimony (MP) above 70% are shown at each node (BI/ML/MP). C. boninense CBS 123755 was used as the outgroup. *Indicates the ex-type strains. Strains isolated in this study are shown in bold.
Taxonomy
Based on morphological traits and multilocus phylogenetic analysis, the 52 isolates were assigned to five Colletotrichum spp. including one new taxon (C. miaoliense sp. nov.) (Fig. 4; described in detail below), one newly recorded taxon in strawberry (C. karstii), and three species known to be associated with strawberry anthracnose (C. boninense, C. fructicola and C. siamense) (Supplementary Fig. S2–S5). The colony features that developed at 25 °C on PDA and 1/4 PDA were all white to grey, with orange conidia ooze. C. siamense ML133 and C. karstii ML351 produced abundant conidia when cultured on 1/4 PDA at 25 °C; C. boninense ML521 produced more conidia on PDA at 25 °C; C. fructicola ML348 and C. miaoliense ML1040 sporulated more abundantly on 1/4 PDA at 30 °C. The conidium and appressorium measurements of the five Colletotrichum spp. (isolates from this study and the type strains) are listed in Supplementary Table S2. The conidia produced by C. miaoliense ML1040 were longer [length to width (L/W) ratio = 3.4] (Fig. 4) than the conidia of the other four species in this study (L/W ratio = 2.3–3) (Fig. S2–S5; Supplementary Table S2).
Colletotrichum miaoliense sp. nov. ML1040. (a) Upper side of colony; (b) reverse side of colony; (c, d) conidiomata; (e–p) appressorium (induced in dH2O on a microscope slide); (q) conidia from conidiomata; (r) conidia from aerial mycelium; (s–w) conidiophores. (a, b) on potato dextrose agar (PDA); (c, d) and (q–w) on 1/4-strength PDA. Scale bars: (c, d) = 0.2 mm, (e) = 10 μm, applies to (f–p, q) = 10 μm, applies to (r–w).
Colletotrichum miaoliense sp. nov. P. C. Chung & H. Y. Wu. Figure 4
MycoBank number MB835424
Etymology: The epithet miaoliense specifically refers to Miaoli County, Taiwan, where the new taxon was discovered.
Sexual morph not observed. Asexual morph observed on 1/4 PDA [BCRC FU31304 (= NTUCC 20-001-1, ML1040)]. Vegetative hyphae 3–6 µm in diameter, hyaline, smooth-walled, septate, branched. Chlamydospores not observed. Sporodochia developed, conidiophores formed directly on hyphae. Conidiophores hyaline, smooth-walled, simple or branched. Conidiogenous cells hyaline, smooth-walled, cylindrical to ampulliform, often integrated, occasionally polyphialidic; phialides discrete, 5.9–26.4 µm (x̅ = 13.3 ± 4.8, n = 55) in length, apical opening 1.1–2.6 µm in diameter (1.7 ± 0.3, n = 55). Conidia hyaline, smooth-walled, aseptate, straight, fusiform to cylindrical, acute ends, 11.2–17 × 3.3–5 µm (x̅ = 14.2 ± 1.1 × 4.1 ± 0.3 µm, n = 100), L/W ratio = 3.4. Conidia from aerial hyphae varied in size (6.6–20 × 2.9–4.9 µm, x̅ = 11.2 ± 2.5 × 3.8 ± 0.4 µm, n = 100), L/W ratio = 3.0. Seta absent. Appressoria single or in loose clusters, pale brown, smooth-walled, elliptical to clavate, entire edge, 5.9–9.1 × 4–6.0 µm (x̅ = 7.5 ± 1.1 × 5 ± 0.6 µm, n = 27), L/W ratio = 1.5.
Culture features: Colonies on PDA flat to somewhat raised, margin entire; mycelium partly floccose, white to pale olivaceous grey; sporodochia orange, scattered in rings, reverse bright orange to orange; average 4.2 cm in diameter in 7 days at 25 °C. Conidia ooze was visible as an orange mass.
Material examined: Taiwan, Miaoli County, Shitan Township, from crown rot of Fragaria × ananassa, 28 Oct. 2016, P.-C. Chung; holotype NTUH 20-001-1, ex-holotype living culture BCRC FU31304 (= NTUCC 20-001-1, ML1040).
Additional materials examined: Taiwan, Nantou County, Renai Township, from leaf spot of Fragaria × ananassa, 24 Nov. 2016, P.-C. Chung, NTUH 20-001-2; living culture NTUCC 20-001-2 (= ML1042). Taiwan, Nantou County, Renai Township, from leaf spot of Fragaria × ananassa, 4 Jul. 2018, P.-C. Chung; NTUH 20-001-3, living culture NTUCC 20-001-3 (= ML1794). Known distribution: Miaoli and Nantou Counties, Taiwan.
Notes Three isolates of C. miaoliense were collected from Miaoli County and Nantou County, Taiwan. Multilocus analysis indicated that C. miaoliense forms a robust clade clearly distinct from all the other known species in the C. acutatum species complex. Of the six Colletotrichum species in this complex (C. acutatum, C. fioriniae, C. godetiae, C. nymphaeae, C. salicis, and C. simmondsii) that have been reported as anthracnose pathogens of strawberry, C. miaoliense is phylogenetically most closely related to C. nymphaeae and C. simmondsii. Morphologically, C. miaoliense differs from C. nymphaeae (CBS 515.78) in the size of conidia (16.1 ± 2.3 × 4.9 ± 0.7 µm versus 14.2 ± 1.1 × 4.1 ± 0.34 µm), the shape of conidia (one end round and one end rounded to acute in contrast to the new species, in which both ends are acute), the size of appressoria (8.7 ± 2.5 × 5.5 ± 1.0 versus 7.5 ± 1. 1 × 5 ± 0.6 µm) (Supplementary Table S2), and the shape of appressoria (C. miaoliense and C. simmondsii appressoria are elliptical to clavate, whereas the appressoria of C. nymphaeae are clavate or irregular in outline, entire, and have an undulate to lobate margin12). Compared with C. simmondsii, the conidia of C. miaoliense are longer (mean length 14.2 µm versus 8.1 µm). In addition, the conidia of C. simmondsii are cylindrical with one end round and one end acute or both ends acute. Although the appressoria of C. miaoliense and C. simmondsii are similar in shape and L/W ratio, the appressoria of C. simmondsii are larger (Supplementary Table S2).
Effect of temperature on mycelial growth
A representative isolate selected from each of five Colletotrichum species was grown on PDA at 18 °C to 32 °C. The maximum growth rate of C. siamense ML133 was estimated at 27.9 °C, whereas the maximum growth rates of C. fructicola ML348, C. karstii ML351, C. boninense ML521 and C. miaoliense ML1040 were at 26.0 °C, 26.9 °C, 24.0 °C, and 26.5 °C, respectively (Fig. 5 and Supplementary Table S3). The growth rate of C. boninense ML521 drastically decreased at 32 °C (Fig. 5). C. miaoliense ML1040 exhibited the slowest growth rate at all tested temperature regimes except 32 °C (Fig. 5). The ranking of species by growth rates at higher temperatures (28 °C, 30 °C and 32 °C) is as follows: C. siamense ML133 > C. fructicola ML348 > C. karstii ML351 and C. boninense ML521 (28 °C and 30 °C) > C. miaoliense ML1040 > C. boninense ML521 (32 °C) (Fig. 5 and Supplementary Table S3).
The growth rates of C. siamense ML133 were also compared with those of another two representative isolates selected from C. siamense, C. fructicola, C. karstii, and C. miaoliense (Supplementary Table S4). The ranking of isolates by mycelial growth rates at both 25 °C and 30 °C on PDA was as follows: C. siamense ML133 and ML540 > C. siamense ML612 and C. fructicola ML368 > C. fructicola ML356 and C. karstii ML1792 > C. karstii ML442 > C. miaoliense ML1042 and ML1794.
Pathogenicity assay
Pathogenicity was tested using Koch's postulates for C. siamense ML133, C. fructicola ML348, C. karstii ML351, C. boninense ML521, and C. miaoliense ML1040. These isolates all caused leaf and/or crown necrosis in strawberry seedlings (Fig. S6). C. siamense ML133 caused the most severe symptoms with 100% disease incidence. The disease incidences for C. fructicola ML348, C. karstii ML351, C. boninense ML521, and C. miaoliense ML1040 were only 30%, 30%, 30% and 50%, respectively. Notably, after spray inoculation of the seedlings with C. karstii ML351, leaf spots scarcely occurred, and no leaf lesions were observed for C. boninense ML521. Even though there were few visible symptoms, C. boninense ML521 could be re-isolated from surface-sterilized inoculated leaves.
Virulence of the five selected isolates was subsequently assayed using wounded and non-wounded detached leaves at 25 °C and 30 °C (Fig. 6). For all five isolates, inoculation of wounded leaves resulted in typical anthracnose lesions, which were first observed at 2–4 days post inoculation (dpi). C. siamense ML133 caused the largest brown necrotic lesions, sometimes with chlorotic or reddish margins (Fig. 6a). The necrotic lesions caused by C. fructicola ML348, C. karstii ML351, C. boninense ML521, and C. miaoliense ML1040 were significantly smaller (Fig. 6b; C. fructicola ML348 was slightly more virulent than C. karstii ML351, C. boninense ML521, and C. miaoliense ML1040). At 7 dpi, C. siamense ML133 caused significantly larger lesions at 30 °C (1.26 cm in diameter) than 25 °C (0.65 cm in diameter), whereas the sizes of lesions caused by other Colletotrichum species were similar (0.07–0.35 cm in diameter) at different temperatures (Fig. 6b).
Inoculation of Colletotrichum spp. on detached wounded and non-wounded strawberry leaves at different temperatures. (a) Lesions at 7 days post inoculation (dpi) on wounded leaves or at 14 dpi on non-wounded leaves incubated at 25 °C or 30 °C. The left side of each leaflet was inoculated with 10 μl spore suspension (106 spores/ml) and the right side with water (control). (b–c) Lesion sizes resulting from inoculation of wounded and non-wounded leaves. The results from the same temperature were analyzed together. Data (mean ± standard error) with different letters are significantly different based on Tukey's range test at P < 0.05 (n = 12). For each species, the difference between 25 °C and 30 °C was analyzed by Student’s t test (*denotes P < 0.05).
In regard to inoculations of unwounded leaves, necrotic lesions caused by C. siamense ML133 and C. fructicola ML348 first appeared at 4–7 dpi, but no lesions occurred in the plants inoculated with the other three Colletotrichum spp. isolates (Fig. 6a). Inoculation of unwounded leaves with C. siamense ML133 resulted in larger lesions than inoculation with the other Colletotrichum spp., and the lesion sizes at 14 dpi were significantly larger at 30 °C (1.35 cm in diameter) than at 25 °C (0.35 cm in diameter) (Fig. 6c).
Inoculations of wounded leaves were conducted at 30 °C for C. siamense ML133, C. boninense ML521, and two additional representative isolates of C. siamense, C. fructicola, C. karstii, and C. miaoliense (Fig. S7). The results showed that C. siamense (ML133, ML540, ML612) and C. fructicola ML356 caused significantly larger lesions (1.21–1.74 cm in diameter at 7 dpi; 3.06–3.47 cm in diameter at 14 dpi) than C. fructicola ML368, C. karstii (ML442, ML1792), and C. miaoliense (ML1042, ML1794) (0.42–0.76 cm in diameter at 7 dpi; 1.06–2.15 cm in diameter at 14 dpi).
Discussion
Over the past decade, our knowledge of fungi and their relationships with plant hosts has seen an exponential growth due to the progress in bioinformatics and molecular phylogenetics. Cryptic taxa identification is progressing rapidly and groups of fungi, including important plant pathogens, are now mainly classified using molecular data-based phylogenetic inference. For instance, the C. acutatum, C. boninense, and C. gloeosporioides species complexes now each contain over 20 species12,13,21. Several Colletotrichum spp. with the capacity to cause strawberry anthracnose in temperate regions have been reported12,13,14,15,17,22,23,24,25,26,27,28,29,30; however, knowledge of the composition of the pathogen populations in tropical and subtropical regions is limited. Through morphological characterization, phylogenetic analyses involving five to seven loci (ITS, GAPDH, CHS-1, ACT, TUB2, CAL, and ApMAT), and inoculation tests on strawberry seedlings and detached leaves, the present study revealed that five Colletotrichum species cause strawberry anthracnose in Taiwan. In addition to the known strawberry anthracnose pathogens C. boninense15, C. fructicola25,31, and C. siamense13,16,18, one new species, C. miaoliense, and one species not previously known to infect strawberry, C. karstii, were identified. C. karstii was previously isolated from a wide range of plants such as anthurium, apple, citrus, and chili pepper21,32,33, but not from strawberry. In this study, C. siamense, C. fructicola, and C miaoliense were isolated from different tissues, and all five Colletotrichum species were proved to be pathogenic to strawberry leaves and crowns (Fig. S6). The lack of tissue specificity is in agreement with previous observations of C. acutatum in strawberry2,34.
The predominance of C. siamense (75%) and C. fructicola (11%) in the strawberry anthracnose pathogen population in Taiwan can be attributed to their higher levels of pathogenicity and aggressiveness. While all five Colletotrichum spp. were pathogenic to wounded leaves, only C. siamense and C. fructicola were able to cause lesions on non-wounded leaves. In addition, C. siamense (ML133, ML540, ML612) and C. fructicola (ML356) caused significantly larger lesions at 25 °C and 30 °C (Fig. 6 and Fig. S7). A similar phenomenon was observed for Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China (latitude ~ 30°N)18: C. fructicola (53% of the isolates) and C. siamense (23%) dominated the population, and C. fructicola exhibited the highest level of pathogenicity (only 25 °C was tested)18. Although C. boninense, C. karstii, and C. miaoliense were much less virulent, wounds caused by natural agencies (wind, rain, insects and animals) as well as human activities (e.g., trimming old leaves, which is a common agricultural practice employed by most strawberry farmers in Taiwan) would provide potential infection sites allowing these pathogens to bypass the first line of defense (e.g., the cuticle)35,36,37 in strawberry.
Among the five Colletotrichum species identified in this study, C. siamense exhibited greater mycelial growth rates on PDA, especially at higher temperatures. The fitness advantage of C. siamense in warm temperature weather may explain its current geographical distribution. In the literature, C. siamense was most reported in tropical and subtropical regions13,16,38, whereas C. boninense, C. fructicola, and C. karstii were reported in regions across a wide range of latitudes13,20,21,33,39,40,41. Temperature is among the key environmental factors affecting a pathogen’s survival42. A recent study based on published observations of 612 crop pests and pathogens from 1960 onwards revealed significant positive latitudinal shifts of many important pests and pathogens under climate change43. More research on the genetic and biological characteristics of different Colletotrichum species from diverse geographical areas will be needed to understand the emergence and spread of anthracnose diseases. With rising global temperatures, it will be particularly important to monitor the expansion of the heat-adapted C. siamense toward high latitudes.
C. boninense, C. fructicola, C. siamense, and C. karstii have been isolated from diverse plants other than strawberry in different countries/regions13,21. In Taiwan, C. fructicola has been reported as an anthracnose pathogen in mango, wax apple, and chili44,45, C. siamense in lychee, star fruit, and mango46,47, C. karstii in passion fruit48, and C. boninense in pitaya49. Previous studies have demonstrated that Colletotrichum spp. from strawberry are pathogenic to other crops and even weeds. For example, C. acutatum could not only infect pepper, eggplant, tomato, and bean but also latently colonize weeds such as Vicia spp. and Conyza spp.50. In one study, C. fructicola was frequently isolated from leaves of Amaranthus blitum, and artificial inoculation of C. fructicola caused brown leaf spots on A. blitum31. To determine whether weed control is necessary to minimize the primary infection in the field, it is worth investigating whether the five Colletotrichum spp. we identified could colonize the weeds commonly present in and nearby strawberry fields in Taiwan. More sampling and artificial inoculation assays will be required to understand the host range of the new species C. miaoliense.
Anthracnose is a key limiting factor for strawberry production in Taiwan and many other areas. Outbreaks of anthracnose in strawberry nurseries and fields have caused yield losses of up to 50–80%2,18,51,52. This study demonstrated the diversity of pathogenic Colletotrichum species associated with strawberry in Taiwan. The findings offer precise information about pathogen identity, which is valuable for screening of resistant varieties and development of effective disease management strategies. Regardless of whether it was inoculated on wounded or non-wounded leaves, the predominant pathogen C. siamense caused larger lesions at 30 °C than 25 °C, which is meaningful in subtropical Taiwan and areas with a similar phenology. Because no significant difference was observed between the mycelial growth rates of C. siamense at 25 °C, 28 °C, and 30 °C, higher disease severity at 30 °C could be due to reduced resistance of strawberry against anthracnose at higher temperatures53,54. In Taiwan, the susceptible cultivar ‘Taoyuan No. 1’ has been widely cultivated for over 30 years. Development of temperature-independent resistant cultivars will be particularly important for strawberry breeding programs in Taiwan and other tropical and subtropical regions. Future work will focus on monitoring pathogen population changes, investigating the fungicide sensitivity levels of different Colletotrichum species, and developing molecular detection methods to aid the production of strawberry seedlings without latent infection of major Colletotrichum species.
Methods
Sample collection and pathogen isolation
From 2010 to 2018, different strawberry tissues (including the leaf, stolon, fruit, root and crown) showing anthracnose disease symptoms were collected from 24 farms and nurseries located in Miaoli, Hsinchu, Nantou, and Chiayi Counties in Taiwan. From 2009 to 2018, the strawberry-cultivated areas in Miaoli, Hsinchu, Nantou, and Chiayi accounted for approximately 89.6%, 2.8%, 2.6%, and 0.2% of the total strawberry-cultivated area in Taiwan, respectively. Pure cultures of all fungal isolates were obtained by the single hyphal tip isolation method. Approximately 2 × 2 mm fragments bordering healthy and necrotic zones in diseased tissues were surface-sterilized with 0.5–1% sodium hypochlorite, rinsed with sterile deionized water three times, then placed onto 1.5% water agar. After 2–3 days of incubation at 25 °C, single hyphal tips were transferred to potato dextrose agar (PDA, BD Difco) and cultured for further use. A total of 52 Colletotrichum spp. isolates were used in this study (Table 1): 26 (50%) isolated from crowns, 11 (21.2%) from leaves, 5 (9.6%) from fruits, 5 (9.6%) from roots, and 5 (9.6%) from stolons (Table 1). Type specimens in this study were deposited in the herbarium of the Department of Plant Pathology and Microbiology, National Taiwan University (NTUH). Ex-type living cultures were deposited in the Culture Collection of the Department of Plant Pathology and Microbiology, National Taiwan University (NTUCC), Bioresource and Collection Research Center (BCRC), and Miaoli District Agricultural Research and Extension Station. Nomenclature and taxonomic information were deposited in MycoBank55 (www.mycobank.org). Colletotrichum spp. were preserved as mycelium discs in ddH2O at 4 °C for short-term storage and in 10% glycerol with 5% lactose at − 80 °C for long term storage. Before conducting experiments, each isolate was revived by culturing on PDA for 5–7 days at 25 °C under a 12-h/12-h photoperiod.
DNA extraction, PCR amplification, and sequencing
For each Colletotrichum spp. isolate, the mycelium was taken from a 7-day-old culture grown on PDA medium. The mycelium was frozen in liquid nitrogen and ground into a fine powder using a sterile mortar and pestle. Genomic DNA was extracted using the Plant Genomic DNA Extraction Miniprep System Kit (VIOGENE) according to the manufacturer’s instructions. Seven genetic fragments, namely ITS, GAPDH, CHS-1, ACT, TUB2, CAL, and ApMAT, were amplified with the primers listed in Supplementary Table S5 using the Biometra Thermal Cycler (Biometra TRIO). Each PCR reaction contained 1 μl of genomic DNA (20–50 ng), 5 μl of 10X reaction buffer [with Tris–HCl (pH 9.0), PCR enhancers, (NH4)2SO4, 20 mM MgCl2], 2 μl of dNTPs (2.5 mM each), 1 µl of 10 μM forward primer, 1 µl of 10 μM reverse primer, 0.5 μl (2.5 U) of Prime Taq DNA Polymerase (GenetBio Inc.), and 39.5 μl of ddH2O. The thermal cycling parameters were 1 cycle of 95 °C for 4 min and 30–35 cycles of 95 °C for 30 s, 52–62 °C for 30 s, and 72 °C for 60 s followed by a final extension step of 72 °C for 7 min. The optimal annealing temperatures for different genetic regions were: ITS: 58 °C, GAPDH: 52 °C, CHS-1: 58 °C, ACT: 58 °C, TUB2: 58 °C, CAL: 59 °C, and ApMAT: 62 °C. Amplicons were bidirectionally sequenced using the dideoxy termination method on the ABI 3730 DNA Analyzer (Tri-I Biotech, Taiwan). Raw sequence chromatograms were manually examined, and the sequences of each fragment were assembled in ApE v2.0.55 (A Plasmid Editor, M. Wayne Davis at the University of Utah, Salt Lake City, UT).
Multilocus phylogenetic analysis and species recognition
Newly generated sequences from each isolate were blasted against the GenBank nr database, and searches were restricted to type materials for initial determination of the closest matching species and species complex. Related gene sequences (ITS, GAPDH, CHS-1, ACT, TUB2, CAL, and ApMAT) of Colletotrichum spp. from recent publications were downloaded from GenBank12,13,21. For each gene, sequences from the isolates belonging to the same species complex were aligned using the MAFFT v7 online server (https://mafft.cbrc.jp/alignment/server/)56. The aligned sequences were manually edited using MEGA v1057 to improve the alignment. The post-alignment sequences of multiple genes/loci were concatenated in a text editor.
BI, ML, and MP approaches for each individual locus and the concatenated sequences were used to identify closely related taxa. Best-fit models of nucleotide substitution were selected using the Akaike information criterion implemented in MrModeltest v.2.458 and run in PAUP v.4.059 (Supplementary Table S6). BI analyses were performed using MrBayes v.3.2.660. Two independent analyses of four Markov Chain Monte Carlo (MCMC) chains (3 heated, 1 cold) were run from a random tree for 2 × 106 (for the C. acutatum species complex), 4 × 106 (for the C. boninense species complex), and 6 × 106 (for the C. gloeosporioides species complex) generations or until the average standard deviation of split frequencies was below 0.01. The analysis was sampled every 1,000 generations, and the first 25% of the generations were discarded as burn-in. The effective sample size and convergence were monitored with Tracer v1.7.161. MP analyses were performed in PAUP v.4.059 using the heuristic search option with Tree Bisection Reconnection branch swapping and 100 random sequence addition. Maxtrees were set to 5,000 and bootstrap analysis was performed with 1,000 replicates. ML analyses were performed in RAxML v8.2.1062 using the GTR-gamma substitution model with 1,000 bootstrap replicates. Phylogenetic trees were visualized in FigTree v1.4.3. The concatenated alignments and phylogenetic trees were deposited in TreeBASE (www.treebase.org) with the study ID 26665.
We applied the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) method63,64 for species delimitation of Colletotrichum taxa. A novel species was considered novel the clade was strongly supported as monophyletic by BI (posterior probability ≥ 0.95), ML (bootstrap ≥ 70%), and MP (bootstrap ≥ 70%) analyses in the multilocus phylogenetic tree and in the majority of individual gene trees.
Morphological characterization
Morphological characterization of one selected isolate from each Colletotrichum species was conducted following the procedures of Weir et al. (2012) and Damm et al. (2012)12,13,21. Cultures were grown on PDA and 1/4-strength PDA (1/4 PDA)65 for 2–3 weeks at 25 °C and 30 °C under a 12-h/12-h photoperiod. (Our preliminary tests showed that compared with PDA, the low nutrient medium 1/4 PDA could stimulate sporulation without affecting the size and shape of conidia.) The experiment was performed in two independent trials, each consisting of two to three plates per isolate. Conidiomata were investigated using a dissecting microscope (Leica M125). Conidia, conidiophores, setae, asci, ascomata and appressoria were examined using a light microscope (Leica DM2500). To induce the formation of appressoria, 20–30 µl of conidial suspension (prepared using sterile dH2O) was dropped onto a microscope slide, covered with a cover slip, then incubated in a moist chamber at 25 °C for 2–3 days39. The lengths and widths of 55 conidiogenous cells, 100 conidia and 30 appressoria were measured using ImageJ software66.
Effect of temperature on mycelial growth
Mycelium-agar discs (6 mm in diameter) were cut with a sterilized cork borer from the advancing edge of 5- to 7-day-old Colletotrichum spp. colonies then placed (with the mycelium-side down) onto the center of a 90 mm petri dish containing 25 ml PDA. Colony diameters were measured after 7 days of incubation at different temperatures under a 12-h/12-h photoperiod in a growth chamber (Firstek, GC-560H). The mycelial growth rate (mm/day) was calculated as “(the diameter of the colony—the diameter of the mycelium-agar disc) / 7”. For one selected isolate from each Colletotrichum species, the mycelial growth rates at 18 °C, 22 °C, 25 °C, 28 °C, 30 °C, and 32 °C were determined in two to three independent trials, each consisting of three PDA plates per isolate per temperature. The optimum temperature for the mycelial growth rate was estimated based on the Gaussian process (4 parameter) for nonlinear regression in SigmaPlot 14 (Systat Software, San Jose, CA). Growth of an additional two representative isolates of each Colletotrichum species, selected from distinct subclades within the species based on the multilocus phylogenetic analysis, was measured at 25 °C and 30 °C in two independent trials, each consisting of four PDA plates per isolate per temperature.
Pathogenicity assay
The susceptible cultivar ‘Taoyuan No. 1’ was used for all inoculation tests in this study. The pathogenicity of Colletotrichum spp. (one isolate from each Colletotrichum species) was examined via Koch's postulates. Strawberry seedlings at the four- to five-leaf stage were inoculated by spraying a spore suspension on the leaves until runoff (106 spores/ml), and also applying 1 ml of spore suspension (106 spores/ml) on the crown region after removal of one to two old leaves. After inoculation, the seedlings were covered with plastic bags (> 90% relative humidity) for 3 days at 30 °C and then incubated in a growth chamber for 11 days at 30 °C and 70% relative humidity under a 12-h/12-h photoperiod. The fungi were re-isolated from lesions of diseased tissues (as mentioned above using the single hypha tip isolation method), then identified based on morphological characteristics and ITS sequences as described above.
The virulence levels of different Colletotrichum spp. isolates were determined by inoculation of wounded and non-wounded detached strawberry leaves. Fully expanded healthy leaves were collected from strawberry seedlings at the four- to five-leaf stage. For inoculation of wounded leaves, each leaflet was punctured with a sterile syringe needle on the left and right sides of the midrib, and 10 μl of a spore suspension (1 × 106 spores/ml) was deposited on the left wound site, and sterile dH2O was deposited on the right wound site as a control. Similarly, inoculations of non-wounded leaves were performed in the same way as mentioned above. After inoculation, the leaves were kept in a moist chamber (a plastic box with dH2O at the bottom; the cut end of the petiole was submerged in the water) at 25 °C or 30 °C under a 12-h/12-h photoperiod. Lesion size was measured at 7 and 14 days post inoculation (dpi). Lesions smaller than 0.1 cm in diameter were considered unsuccessful infections. The same isolates for the “effect of temperature on mycelial growth” test were used for the pathogenicity assay. For one selected isolate from each Colletotrichum species, inoculations of wounded and non-wounded leaves were performed at 25 °C and 30 °C in three independent trials, each consisting of 12 leaflets (4 leaves from 4 seedlings) per isolate per treatment. An additional two representative isolates of each Colletotrichum species, selected from distinct subclades within the species based on the multilocus phylogenetic analysis, were used for wound inoculation at 30 °C. The experiment was performed in two independent trials, each consisting of 12 leaflets (4 leaves from 4 seedlings) per isolate per treatment.
Statistical analysis
Data were analyzed by analysis of variance (ANOVA) using the software SPSS v18. Tukey’s range test or Student’s t-test was used to test for significant differences among or between different treatments at a significance threshold of P < 0.05.
References
FAOSTAT. Food and Agriculture Organization Statistical Database. Groundnut production and export data. https://www.fao.org/faostat/en/#data/QC (Accessed 20 April 2020). (2020).
Howard, C. M., Maas, J. L., Chandler, C. K. & Albregts, E. E. Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Dis. 76, 976–981 (1992).
Penet, L., Guyader, S., Petro, D., Salles, M. & Busslere, F. Direct splash dispersal prevails over indirect and subsequent spread during rains in Colletotrichum gloeosporioides infecting yams. PLoS ONE 9, e115757 (2014).
Perfect, S. E., Hughes, H. B., O’Connell, R. J. & Green, J. R. Colletotrichum—a model genus for studies on pathology and fungal-plant interactions. Fungal Genet. Biol. 27, 186–198 (1999).
Chung, P. C., Wu, H. Y., Ariyawansa, H. A., Tzean, S. S. & Chung, C. L. First report of anthracnose crown rot of strawberry caused by Colletotrichum siamense in Taiwan. Plant Dis. 103, 1775–1776 (2019).
Gunnell, P. S. & Gubler, W. D. Taxonomy and morphology of Colletotrichum species pathogenic to strawberry. Mycologia 84, 157–165 (1992).
Cannon, P. F., Damm, U., Johnston, P. R. & Weir, B. S. Colletotrichum—current status and future directions. Stud. Mycol. 73, 181–213 (2012).
Freeman, S. & Katan, T. Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathology 87, 516–521 (1997).
Cai, L. et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 39, 183–204 (2009).
Hyde, K. D. et al. Colletotrichum: a catalogue of confusion. Fungal Divers. 39, 1–17 (2009).
Vieira, W. A. S. et al. The impact of phenotypic and molecular data on the inference of Colletotrichum diversity associated with Musa. Mycologia 109, 912–934 (2017).
Damm, U., Cannon, P. F., Woudenberg, J. H. C. & Crous, P. W. The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113 (2012).
Weir, B. S., Johnston, P. R. & Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180 (2012).
Baroncelli, R. et al. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum Species. PLoS ONE 10, e0129140 (2015).
Bi, Y., Guo, W., Zhang, G. J., Liu, S. C. & Yang, B. D. First report of Colletotrichum boninense causing anthracnose of strawberry in China. Plant Dis. 101, 250–251 (2017).
Han, Y. C. et al. Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei, Chinai. Plant Dis. 100, 996–1006 (2016).
Jayawardena, R. S. et al. An account of Colletotrichum species associated with strawberry anthracnose in China based on morphology and molecular data. Mycosphere 7, 1147–1163 (2016).
Chen, X. Y. et al. Genetic diversity of Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China. Plant Dis 104, 1351–1357 (2020).
Tzean, S. S. et al. List of Plant Diseases in Taiwan 5th edn, 140–141 (Taiwan Phytopathological Society Press, Taiwan, 2019).
Fu, M. et al. Colletotrichum species associated with anthracnose of Pyrus spp. China. Persoonia 42, 1–35 (2019).
Damm, U. et al. The Colletotrichum boninense species complex. Stud. Mycol. 73, 1–36 (2012).
Braganca, C. A. D., Damm, U., Baroncelli, R., Massola, N. S. & Crous, P. W. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biol. 120, 547–561 (2016).
Porta-Puglia, A. & Mifsud, D. First record of Colletotrichum acutatum strawberry in Malta. J. Plant Pathol. 88, 228–228 (2006).
Adhikari, T. B., Chacon, J. G., Fernandez, G. E. & Louws, F. J. First report of anthracnose causing both crown and fruit rot of strawberry by Colletotrichum siamense in North Carolina. Plant Dis. 103, 1775–1775 (2019).
Gan, P., Nakata, N., Suzuki, T. & Shirasu, K. Markers to differentiate species of anthracnose fungi identify Colletotrichum fructicola as the predominant virulent species in strawberry plants in Chiba Prefecture of Japan. J. Gen. Plant. Pathol. 83, 14–22 (2017).
Nam, M. H., Kim, T. I., Gleason, M. L., Song, J. Y. & Kim, H. G. First report of anthracnose fruit rot caused by Colletotrichum acutatum on strawberry in Korea. Plant Dis. 92, 1247–1247 (2008).
Latinovic, J., Latinovic, N., Tiodorovic, J. & Odalovic, A. First report of anthracnose fruit rot of strawberry caused by Colletotrichum acutatum in Montenegro. Plant Dis. 96, 1066–1066 (2012).
Polizzi, G. et al. First report of damping-off on strawberry tree caused by Colletotrichum acutatum and C. simmondsii in Italy. Plant Dis. 95, 1588–1589 (2011).
Bobev, S. G., Zveibil, A. & Freeman, S. First report of Colletotrichum acutatum on strawberry in Bulgaria. Plant Dis. 86, 1178–1178 (2002).
Novotny, D., Krizkova, I. & Salava, J. First report of anthracnose caused by Colletotrichum acutatum on strawberry in the Czech republic. Plant Dis. 91, 1516–1516 (2007).
Hirayama, Y., Asano, S., Okayama, K., Ohki, S. T. & Tojo, M. Weeds as the potential inoculum source of Colletotrichum fructicola responsible for strawberry anthracnose in Nara Japan. J. Gen. Plant. Pathol. 84, 12–19 (2018).
Diao, Y. Z. et al. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 38, 20–37 (2017).
Guarnaccia, V., Groenewald, J. Z., Polizzi, G. & Crous, P. W. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 39, 32–50 (2017).
Peres, N. A., Timmer, L. W., Adaskaveg, J. E. & Correll, J. C. Lifestyles of Colletotrichum acutatum. Plant Dis. 89, 784–796 (2005).
Ranathunge, N. P., Mongkolporn, O., Ford, R. & Taylor, P. W. J. Colletotrichum truncatum pathosystem on Capsicum spp: infection, colonization and defence mechanisms. Australas. Plant Path. 41, 463–473 (2012).
Sm Auyong, A. The role of cutinase and its impact on pathogenicity of Colletotrichum truncatum. J. Plant Pathol. Microbiol. 6, 259–270 (2015).
Wang, Y. X., Chen, J. Y., Li, D. W., Zheng, L. & Huang, J. B. CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of Camellia oleifera. Eur. J. Plant Pathol. 147, 103–114 (2017).
Silva, D. N. et al. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia 104, 396–409 (2012).
Yang, Y. L. et al. Colletotrichum anthracnose of Amaryllidaceae. Fungal Divers. 39, 123–146 (2009).
Hu, M. J., Grabke, A. & Schnabel, G. Investigation of the Colletotrichum gloeosporioides species complex causing peach anthracnose in south Carolina. Plant Dis. 99, 797–805 (2015).
Li, Q. et al. Colletotrichum species associated with mango in southern China. Sci. Rep. 9, 18891 (2019).
Velásquez, A. C., Castroverde, C. D. M. & He, S. Y. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 28, 619–634 (2018).
Bebber, D. P., Ramotowski, M. A. T. & Gurr, S. J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change. 3, 985–988 (2013).
Duan, C. H., Pan, H. R. & Wang, C. C. Identification, pathogenicity and fungicide sensitivity of Colletotrichum isolates from five fruit crops in Taiwan. Taiwan Pestic Sci. 5, 91–111 (2018).
de Silva, D. D. et al. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 10, 8 (2019).
Ni, H. F. et al. First report of pepper spot disease of lychee caused by Colletotrichum siamense in Taiwan. J. Plant Pathol. 99, 808–808 (2017).
Wu, C. J., Chen, H. K. & Ni, H. F. Identification and characterization of Colletotrichum species associated with mango anthracnose in Taiwan. Eur. J. Plant Pathol. 157, 1–15 (2020).
Lin, Y. H. Identification of Antagonistic Microorganisms Against Anthracnose on Tea and Passion Fruit and Their Potential Application In Disease Control (National Chung Hsing University, Taichung City, 2015).
Lin, C. P., Ann, P. J., Huang, H. C., Chang, J. T. & Tsai, J. N. Anthracnose of pitaya (Hylocereus spp.) caused by Colletotrichum spp., a new postharvest disease in Taiwan. J. Taiwan Agric. Res. 66, 171–183 (2017).
Freeman, S., Horowitz, S. & Sharon, A. Pathogenic and nonpathogenic lifestyles in Colletotrichum acutatum from strawberry and other plants. Phytopathology 91, 986–992 (2001).
Forcelini, B. B. & Peres, N. A. Widespread resistance to QoI fungicides of Colletotrichum acutatum from strawberry nurseries and production fields. Plant Health Prog. 19, 338–341 (2018).
Xie, L., Zhang, J. Z., Wan, Y. & Hu, D. W. Identification of Colletotrichum spp. isolated from strawberry in Zhejiang Province and Shanghai City. China. J. Zhejiang Univ. Sci. B 11, 61–70 (2010).
Mori, T. Effects of temperature as the selection pressure for resistance to anthracnose crown rot (Glomerella cingulata Spaulding et Schrenk) of young strawberry seedlings. J. Jpn. Soc. Hortic. Sci. 67, 934–938 (1998).
Smith, B. J. & Black, L. L. Resistance of strawberry plants to Colletotrichum fragariae affected by environmental conditions. Plant Dis. 71, 834–837 (1987).
Crous, P., Stalpers, J. & Robert, V. MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22 (2004).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Nylander, J. MrModeltest V2. Program distributed by the author. 24 (2004).
L. Swofford, D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10. Vol. Version 4.0 (2002).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9), 1312–1313 (2014).
Dettman, J. R., Jacobson, D. J. & Taylor, J. W. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57, 2703–2720 (2003).
Taylor, J. W. et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31, 21–32 (2000).
Su, Y.-Y., Qi, Y.-L. & Cai, L. Induction of sporulation in plant pathogenic fungi. Mycology 3, 195–200 (2012).
Rueden, C. T. et al. Image J2: ImageJ for the next generation of scientific image data. BMC Bioinform. 18, 529 (2017).
Acknowledgements
We acknowledge Dr. Dai-Rong Wu for sharing plant materials. We thank Ms. Feng-Ching Tu, Shih-Jhu Jiang, and Mr. Ji-Feng Li (Miaoli District Agricultural Research and Extension Station, Taiwan) for help in the inoculation trials. This research was funded by the Ministry of Science and Technology, Taiwan (grant no. 108-2321-B-002-023-).
Author information
Authors and Affiliations
Contributions
P.C.C., H.Y.W., T.H.H., S.S.T. and C.L.C. designed the experiments, analyzed the data, and drafted the manuscript. P.C.C., H.Y.W., Y.W.W., and H.P.H. performed the experiments. P.C.C., H.Y.W. and H.A.A. conducted phylogenetic analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chung, PC., Wu, HY., Wang, YW. et al. Diversity and pathogenicity of Colletotrichum species causing strawberry anthracnose in Taiwan and description of a new species, Colletotrichum miaoliense sp. nov.. Sci Rep 10, 14664 (2020). https://doi.org/10.1038/s41598-020-70878-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70878-2
This article is cited by
-
Identification, prevalence and pathogenicity of Colletotrichum species associated with chilli anthracnose in India
Journal of Plant Pathology (2023)
-
Grape ripe rot disease caused by Colletotrichum spp.: current status, concerns and future perspectives
Journal of Plant Pathology (2023)
-
Colletotrichum species causing anthracnose disease on avocado fruit in Taiwan
European Journal of Plant Pathology (2023)
-
Different responses to elevated temperature in the representative strains of strawberry pathogenic Colletotrichum spp.from eastern China
Mycological Progress (2023)
-
Colletotrichum species pathogenic to strawberry: discovery history, global diversity, prevalence in China, and the host range of top two species
Phytopathology Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.